Abstract
Microcotyle sebastis is a gill monogenean ectoparasite that causes serious problems in the mariculture of the Korean rockfish, Sebastes schlegelii. In this study, we isolated the parasite from fish farms along the coasts of Tongyeong, South Korea in 2016, and characterized its infection, morphology and molecular phylogeny. The prevalence of M. sebastis infection during the study period ranged from 46.7% to 96.7%, and the mean intensity was 2.3 to 31.4 ind./fish, indicating that the fish was constantly exposed to parasitic infections throughout the year. Morphological observations under light and scanning electron microscopes of the M. sebastis isolates in this study showed the typical characteristics of the anterior prohaptor and posterior opisthaptor of monogenean parasites. In phylogenetic trees reconstructed using the nuclear 28S ribosomal RNA gene and the mitochondrial cytochrome c oxidase I gene (cox1), they consistently clustered together with their congeneric species, and showed the closest phylogenetic relationships to M. caudata and M. kasago in the cox1 tree.
-
Key words: Microcotyle sebastis, fish farm, Korean rockfish, monogenean parasite, infection, molecular phylogeny, morphology
The Korean rockfish,
Sebastes schlegelii, is an ovoviviparous fish that inhabits the coastal areas of China, Japan, and Korea. It is one of the most important fisheries resources in South Korea, and its mariculture has increased rapidly over the past decades [
1]. However, one problem associated with the mariculture of the fish is infections with a monogenean parasite,
Microcotyle sebastis on its gills, which causes extensive mortality in the juveniles every year [
1].
Monogeneans are ectoparasites on the body and gills of freshwater and marine fishes, exhibiting high host specificity. They are sometimes hyperparasitic and cause serious problems in fish aquaculture worldwide [
2]. Although they have not been reported to be overtly pathogenic, their heavy infection in individual hosts can be pathogenic and cause extensive mortality [
3].
The genus
Microcotyle Van Beneden & Hesse, 1863 is a flatworm belonging to the class Monogenea. It is one of the oldest monogenean genera, including many species [
4]. This genus is notorious for causing difficulties in taxonomy, and several attempts have been made to subdivide it into separate genera [
5,
6]. Thus, integrative studies based on morphology and molecular phylogeny are necessary to elucidate its phylogenetic relationships [
5]. For example, Víllora-Montero et al. [
7] redescribed
Microcotyle erythrini van Beneden & Hesse, 1863, originally reported as
Pagellus erythrinus (L.), based on molecular data, defined its morphological boundaries, and provided new morphological and molecular data for
Microcotyle species identification. They recommended using molecular markers such as the nuclear 28S ribosomal RNA gene (28S rDNA) and the mitochondrial cytochrome
c oxidase I gene (
cox1) as complementary information for interspecific differentiation. The mitochondrial
cox1 was recently used to identify and reconstruct phylogenetic trees among vertebrate parasites [
8]. Comparative analysis of their mitochondrial genomes, involving features such as gene arrangement, nucleotide sequences, and amino acid sequences, has become a powerful molecular approach to resolving the deeper nodes of phylogenetic relationships in a variety of metazoan groups [
9].
Several studies have been conducted on
M. sebastis infections in the Korean rockfish in South Korea, including surveys of their seasonal prevalence [
10], egg production, embryonic development [
11], chemotherapeutics [
12], and mitochondrial genome [
13]. However, there is a lack of information on infection at fish farms, morphological characteristics, and molecular data. In this study, we investigated molecular phylogeny based on nuclear 28S rDNA and mitochondrial
cox1 sequence data, morphological observation, and infection status of
M. sebastis at fish farms along the coasts of Tongyeong, Korea.
Thirty specimens of the Korean rockfish were sampled from each sampling station (net cage fish farm). Thus, a total of 270 specimens were collected from 9 stations along the coasts of Tongyeong, South Korea, in 2016 (
Table 1); 3 stations in March (group 1), 3 stations in June and July (group 2), 3 stations in October (group 3). They were transported to the laboratory in a polythene bag with ice. One side of the operculum was removed, and 4 gill arches were sequentially excised and soaked in a Petri dish containing sterile phosphate-buffered saline (PBS) buffer to prevent drying. The number of
M. sebastis from each fish specimen was counted under a stereomicroscope, SMZ-171 (Motic, Hong Kong, China). The prevalence was defined as the proportion of infected hosts among all hosts examined, and the mean intensity was calculated as the mean number of parasites found in each infected host, excluding zero values of uninfected hosts. Differences in the mean prevalence among the 3 groups were tested by a one-way ANOVA test using GraphPad InStat (GraphPad Software Inc., San Diego, California, USA) and considered significant at
P<0.05.
For morphometric analysis, M. sebastis was isolated using tweezers and observed under a stereomicroscope after fixing in 10% formalin. For scanning electron microscope (SEM) observation, the isolates were washed twice with sterile seawater and fixed with 1% OsO4 (Sigma-Aldrich, St. Louis, Missouri, USA) for 10 min. Thereafter, they were rinsed twice with distilled water, placed on a Nuclepore™ polycarbonate membrane filter (Corning, New York, New York, USA), dehydrated in a graded ethanol series, and finally substituted with isoamyl alcohol. The fixed isolates were dried in a critical point dryer (K850 Critical Point Dryer, Pelco, Germany) using liquid CO2 for 3 hr, coated with Au-Pd in Ion Sputter (E-1000, Hitachi, Tokyo, Japan), and examined under JSM-6490LV Scanning Electron Microscope (JEOL, Tokyo, Japan).
For the DNA sequence analyses, the 5 isolates of
M. sebastis were individually isolated under a stereomicroscope and placed into sterile microtubes. Their genomic DNA (gDNA) was extracted using DNeasy
® Tissue&Blood Kits (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Independent PCR amplifications were conducted for the nuclear 28S rDNA and mitochondrial
cox1 in a 20 μl reaction volume containing 1 μl of gDNA, 1×
Ex Taq Buffer, 0.2 μM of the forward and reverse primers, 250 μM dNTP mix, and 0.1 units of
TaKaRa Ex Taq™ (TaKaRa Bio Inc., Shiga, Japan). The PCR primers for nuclear 28S rDNA were based on those described by Scholin et al. [
14] and Nunn et al. [
15], and those for mitochondrial
cox1 were newly designed for this study: Monog-CO1f, 5′-CTGTTACTGCTAATAATGGT-3′, Monog-CO1r, 5′-GCTAATCATGATGCAAAAGG-3′. PCR amplifications were performed using the following thermal cycling profile in a ProFlex PCR System (Thermo Fisher Scientific Inc., Waltham, Massachusetts, USA): initial denaturation at 94°C for 1 min, 35 cycles of denaturation at 98°C for 10 sec, annealing at 55°C for 30 sec, and elongation at 72°C for 2 min. The reaction was completed with a final elongation step at 72°C for 10 min. The PCR products were purified using an
AccuPrep® PCR Purification Kit (Bioneer, Daejeon, South Korea) and directly sequenced on an Applied Biosystems
® 3730
xl DNA Analyzer (Applied Biosystems, Carlsbad, California, USA) with the primers used in the PCR amplifications. The sequences of the nuclear 28S rDNA and mitochondrial
cox1 analyzed in this study were deposited at a public sequence database, GenBank of the National Center for Biotechnology Information (NCBI) (
https://www.ncbi.nlm.nih.gov/) under accession numbers MT875155-MT875159 and MT876115-MT876119, respectively.
The nuclear 28S rDNA and mitochondrial
cox1 sequences of all species belonging to the family Microcotylidae were retrieved from GenBank. They were aligned with the
M. sebastis sequences analyzed in this study, using ClustalW in BioEdit 7.2 [
16] and refined manually to correct obvious misalignments. The final sequence matrices consisting of 799 bp and 963 bp, respectively, were subjected to maximum likelihood (ML) analyses using MEGA 7 [
17]. The best-fit substitution models were selected using MEGA 7 for the ML analyses, and Tamura-Nei (TN93) and Gamma distribution (G) were applied for both genes. Statistical support for each node was evaluated using bootstrapping with 1,000 replicates. In addition, Bayesian inference (BI) analyses were conducted with generalized time-reversible (GTR) and G model using MrBayes 3.1.2 [
18] for both genes. Two independent Metropolis-coupled Markov chain Monte Carlo (MCMCMC) runs were launched with 4 chains each, 3 heated and 1 cold, and random starting trees for 5,000,000 generations and sampling the trees at intervals of 100 generations. A total of 10,000 of the 50,001 resulting trees were discarded as “burn-in”. The remaining trees were used to reconstruct a 50% majority-rule consensus tree, and TreeView 1.6.6 [
19] was used to summarize the posterior probability support for each node.
Previous studies have reported seasonal changes in the prevalence and mean intensity of monogenean parasites, relating to water temperature, host immunological resistance and host ages [
10,
20]. Yoon et al. [
10] reported winter and summer peaks of
M. sebastis infection in the Korean rockfish, and Ogawa [
20] observed winter and spring peaks of
Bivagina tai in the red seabream,
Pagrus major. In the present study, the mean prevalence of
M. sebastis among the 3 groups of the Korean rockfish was significantly different (
P=0.014), with the highest prevalence in G1, followed by G3 and G2. In addition, its mean intensity was also higher in G1 (8.5–31.4) than in G2 (4.1–5.5) and G3 (2.3–3.5). From these results, it seems to show that the infection was peak in spring (G1) in this study. However, chemotherapeutic drugs such as praziquantel and trichlorfon are commonly used to treat the parasitic infection in aquaculture, and it could affect the prevalence or intensity of the infection as well as water temperature and host immunity. Since this study did not investigate whether chemotherapy was used or not when we sampled the fish, it was insufficient to conclude the spring peak of the infection despite the significantly high prevalence and intensity of the infection in G1. Therefore, in the present study, it was only confirmed that the fish were continuously exposed to parasitic infection throughout the year.
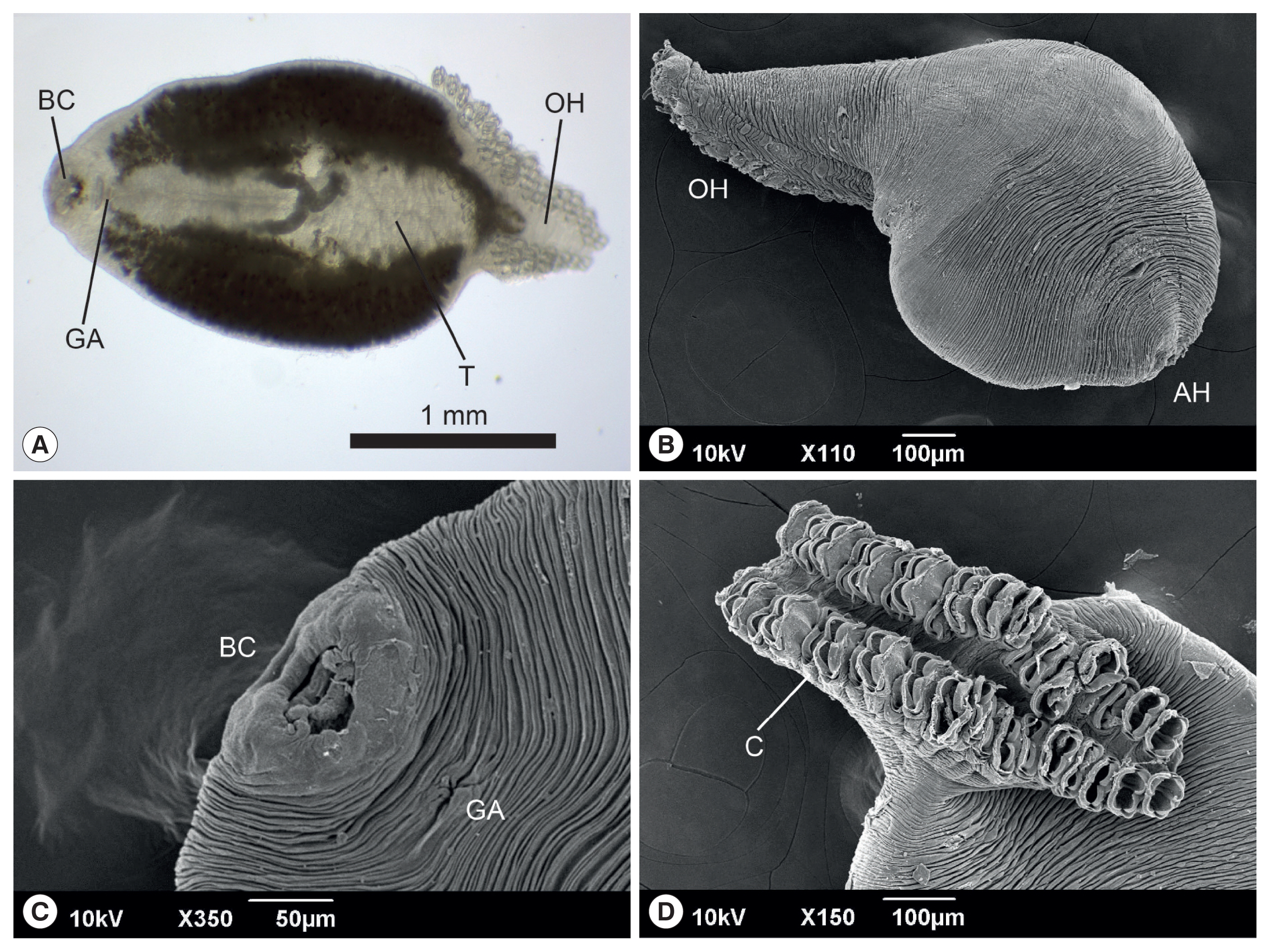
The anterior end of the body of
M. sebastis, called the prohaptor, has various feeding and adhesive structures. They also possess a posterior attachment organ, called the opisthaptor or haptor. However, few reports on its morphological characteristics exist, except for metric analyses [
21,
22]. In the present study, we produced microphotographs of the whole body, prohaptor, and opisthaptor of
M. sebastis, typical morphological characteristics of flatworms in the subclass Polyopisthocotylea, using light microscope and SEM. Its body was round and flat (
Fig. 1A, B) with an anterior prohaptor (
Fig. 1C) and a posterior opisthaptor (
Fig. 1D). The buccal cavity was located in the anterior part of the ventral surface. The results of the morphometric analysis was as follows (n>20): body length, 2,052±597 μm (1,502–3,055 μm); body width, 965±165 μm (680–1,245 μm); posterior opisthaptor length, 855±592 μm; clamp length, 73±50 μm; clamp width, 43±30 μm; genital atrium length, 115±71 μm; genital atrium width, 52±34 μm; number of clamp, 42–48; and number of testes, 21–25.
M. sebastis in the present study was morphologically similar to
M. kasago and
M. caudata, which were isolated from the host species belonging to the subfamily Sebastinae [
23]. However, the morphometric data showed slight differences among the 3 species; their average body length was similar (
M. caudata 2,769 μm;
M. kasago 2,669 μm), with the largest size in
M. caudata (5,550 μm); the number of clamps was in the range of 27–69 in
M. caudata and 32–43 in
M. kasago; and the number of testes was in the range of 4–24 in
M. caudata and 10–14 in
M. kasago. The SEM observation showed that the body surface was covered with long and deep wrinkles and the opisthaptor, arranged in 2 rows, was separated from the actual body at the posterior region and composed of 2 symmetrical rows of clamps, formed by 2 opposable hinged jaws.
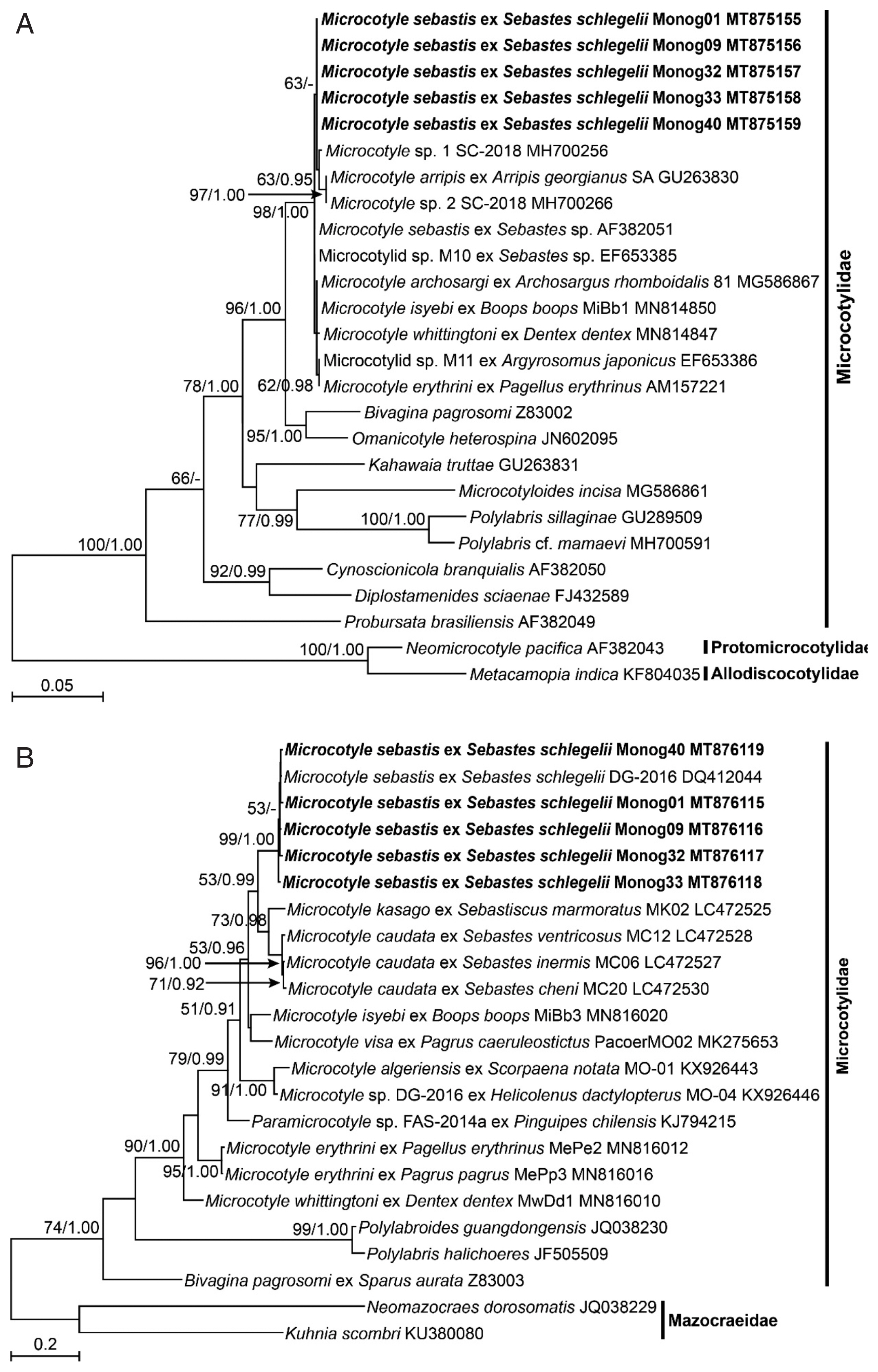
The DNA sequences of the nuclear 28S rDNA and mitochondrial
cox1 of the 5 isolates of
M. sebastis were subjected to molecular phylogenetic analyses. They had identical lengths of 791 bp and 963 bp, respectively, without insertions or deletions, and showed similarities of 100% and 98.5–99.0% to each other, respectively. The 28S rDNA sequences showed the highest similarity with
M. sebastis (GenBank accession number AF382051) and Microcotylidae sp. M10 (EF653385) (99.87%) in BLAST search, and the
cox1 sequences did with
M. sebastis (DQ412044) (98.86–99.38%). In the ML and BI trees inferred from both genes, all species belonging to the genus
Microcotyle consistently clustered together with the highest statistical supports (
Fig. 2A, B). In the nuclear 28S rDNA tree (
Fig. 2A), the 5 isolates in this study consistently clustered with 6
Microcotyle species, including
M. sebastis, but no clear phylogenetic separations were found among the species because of low statistical supports. In contrast, in the mitochondrial
cox1 tree (
Fig. 2B), most nominal species were clearly separated, except for the branch between
M. visa and
M. isyebi. The 5 isolates showed the closest phylogenetic relationships with
M. caudata and
M. kasago. The
M. sebastis isolates from the host species, the Korean rockfish, in this study showed the closest phylogenetic relationships with
M. caudata, which has
Sebastis cheni,
S. inermis, or
S. ventricosus as host species, and with
M. kasago, which has
Sebasticus mamoratus as a host species. All of the host species belong to the subfamily Sebastinae. The 2 host genera share a unique fertilization pattern, viviparity, and identical morphological characteristics [
24]. Thus, our results revealed that phylogenetic analysis using mitochondrial
cox1 sequences is well congruent with the host specificity of the monogenean parasites at the subfamilial level.
Meanwhile, our phylogenetic analyses of the
Microcotyle species based on nuclear 28S rDNA showed low phylogenetic resolution, but high resolution based on mitochondrial
cox1, of which the result is in agreement with previous studies [
7,
25]. Mitochondrial
cox1 is a well-known molecular marker for metazoan DNA barcoding, because of its intra- and interspecific variation and adequate barcode gap [
26]. This marker is also a gold standard for studies on population genetics, host specificity range, and the phylogenetics and systematics of monogenean parasites.
In this study, we investigated the molecular phylogeny of a monogenean parasite, M. sebastis, along with its morphological observations and infection status in the Korean rockfish. This parasite can cause loss of stock in the Koran rockfish, but to date, it has not been extensively studied. Our results will provide the information on more efficient management of fishery stocks, morphological identification, and phylogenetic analysis of the parasite.
Notes
-
CONFLICT OF INTEREST
The authors declare no conflict of interest related to this study.
ACKNOWLEDGMENT
This work was supported by a grant from the National Institute of Fisheries Science, Republic of Korea (R2021065).
Fig. 1Light photomicrograph (A) and scanning electron microphotographs (B–D) of Microcotyle sebastis isolated from the Korean rockfish, Sebastes schlegelii. AH, anterior prohaptor; BC, buccal cavity; C, clamp; GA, genital atrium; OH, opisthaptor; T, testes.
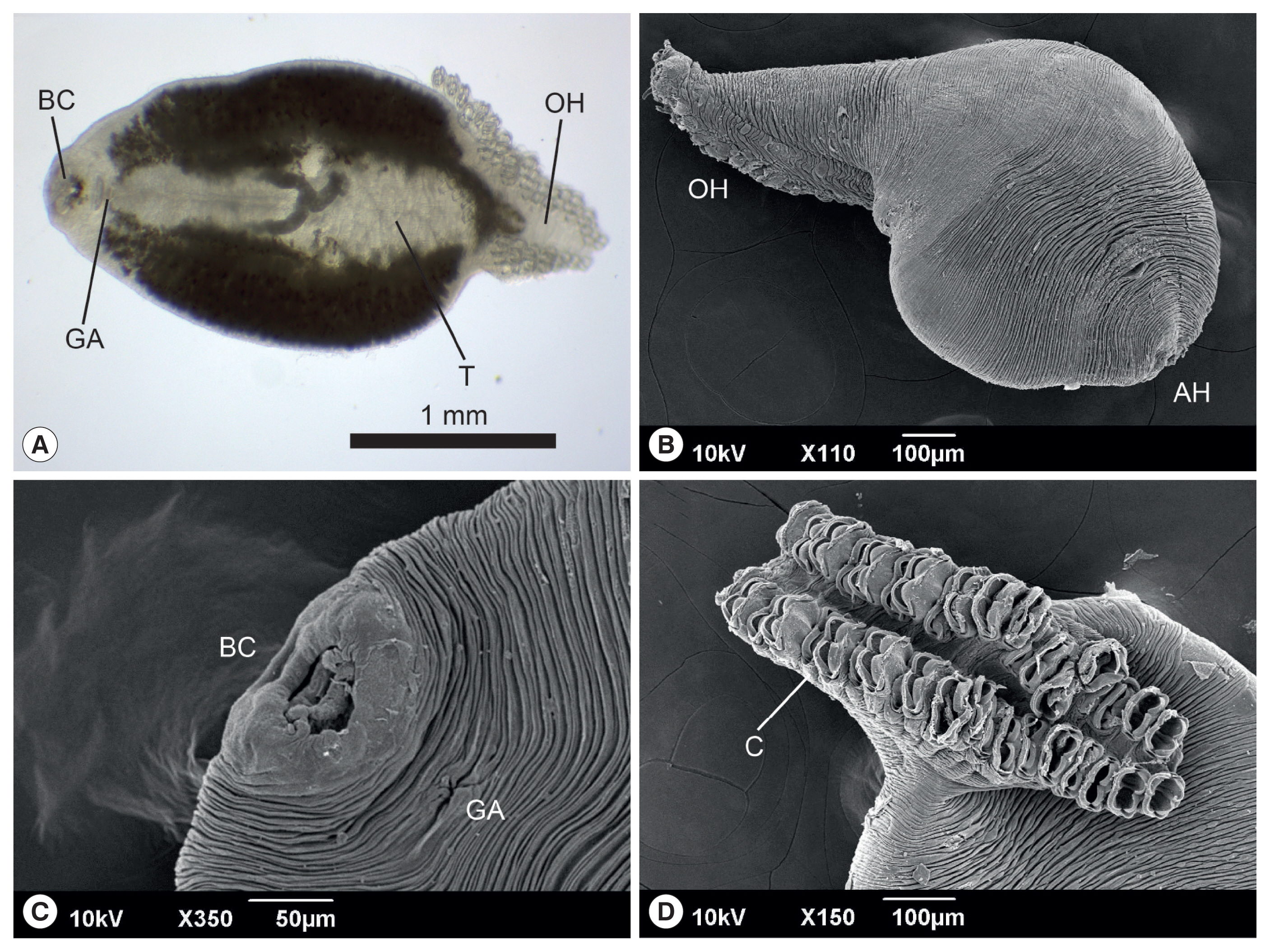
Fig. 2Maximum likelihood (ML) tree of monogenean parasites belonging to the family Microcotylidae, based on a sequence matrix of nuclear 28S ribosomal RNA gene (A) and mitochondrial cytochrome c oxidase I gene (B). Numbers at the branch nodes indicate the bootstrapping values obtained in a ML analysis, and the posterior probability in a Bayesian analysis. The scale bar indicates the estimated number of nucleotide substitutions per site. The species newly analyzed in this study is shown in bold face. The Microcotyle species are indicated with their host species.
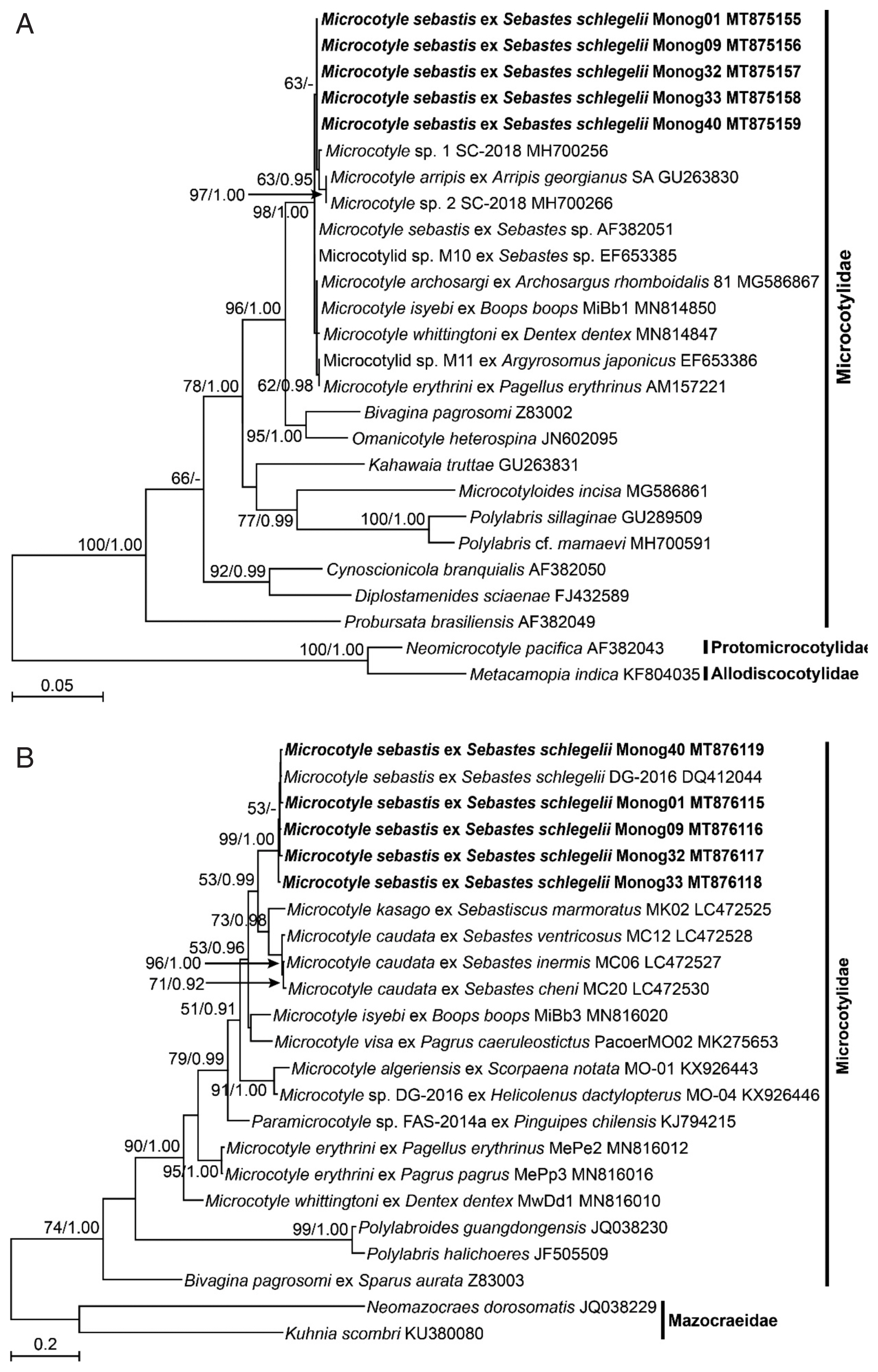
Table 1Prevalence and intensity of the monogenean parasite, Microcotyle sebastis to the Korean rockfish, Sebastes schlegelii at the fish farms along the coasts of Tongyeong, Korea in 2016
Table 1
|
Group |
Station (fish farm) |
Sampling month |
No. of tested fish |
Mean length±SD (cm) |
Mean weigh±SD (g) |
Prevalence (no. of positive fish) |
Mean prevalence±SD |
No. of parasite/fish (mean±SD) |
|
G1 |
St. 1 |
Mar. |
30 |
17.7±1.2 |
100.9±20.1 |
90.0 (27) |
93.3±0.03 |
2–19 (8.5±4.8) |
|
St. 2 |
Mar. |
30 |
23.3±2.1 |
206.1±63.8 |
96.7 (29) |
1–34 (12.7±10.0) |
|
St. 3 |
Mar. |
30 |
20.0±0.9 |
172.4±23.8 |
93.3 (28) |
1–68 (31.4±15.8) |
|
|
G2 |
St. 4 |
Jun. |
30 |
20.2±8.26 |
187.5±206.6 |
60.0 (18) |
60.0±0.13 |
1–13 (4.1±3.35) |
|
St. 5 |
Jul. |
30 |
27.1±2.3 |
319.1±79.7 |
46.7 (14) |
1–19 (4.6±5.12) |
|
St. 6 |
Jul. |
30 |
11.8±1.2 |
24.8±9.4 |
73.3 (22) |
1–13 (5.5±3.6) |
|
|
G3 |
St. 7 |
Oct. |
30 |
16.5±2.5 |
65.71±28.3 |
86.7 (26) |
76.7±0.09 |
1–7 (3.5±2.0) |
|
St. 8 |
Oct. |
30 |
16.8±1.6 |
71.5±21.3 |
73.3 (22) |
1–7 (2.7±1,6) |
|
St. 9 |
Oct. |
30 |
14.4±1.6 |
41.3±15.0 |
70.0 (21) |
1–7 (2.3±1.7) |
References
- 1. Kim CS, Cho JB, Ahn KJ, Lee JI, Kim KH. Treatment of Microcotyle sebastis (Monogenean: Polyopisthocotylea) infestation with praziquantel under commercial rockfish (Sebastes schlegeli) culture condition. J Fish Sci Tech 2002;5:141-143.
https://doi.org/10.5657/fas.2002.5.2.141
- 2. Rhode K. Diseases caused by metazoans: Helminths. In Kinne O ed, Diseases of marine animals, Vol IV, Part 1, Introduction, Pisces. Hamburg, Germany. Biologische Anstalt Helgoland; 1984, pp 193-320.
- 3. Thoney DA. Post-larval growth of Microcotyle sebastis (Platyhelminthes: Monogenea), a gill parasite of the black rockfish. Trans Am Microsc Soc 1986;105:170-181.
https://doi.org/10.2307/3226389
- 4. Machkewskyi VK, Dmitrieva EV, Al-Jufaili S, Al-Mazrooei NA.
Microcotyle omanae n. sp. (Monogenea: Microcotylidae), a parasite of Cheimerius nufar (Valenciennes) (Sparidae) from the Arabian Sea. Syst Parasitol 2013;86:153-163.
https://doi.org/10.1007/s11230-013-9444-5
- 5. Bouguerche C, Gey D, Justine JL, Tazerouti F.
Microcotyle visa n. sp. (Monogenea: Microcotylidae), a gill parasite of Pagrus caeruleostictus (Valenciennes) (Teleostei: Sparidae) off the Algerian coast, Western Mediterranean. Syst Parasitol 2019;96:131-147.
https://doi.org/10.1007/s11230-019-09842-2
- 6. Mamaev Yl. The taxonomical composition of the family Microcotylidae Taschenberg, 1879 (Monogenea). Folia Parasitol 1986;33:199-206.
- 7. Víllora-Montero M, Pérez-del-Olmo A, Georgieva S, Raga JA, Montero FE. Considerations on the taxonomy and morphology of Microcotyle spp.: redescription of M. erythrini van Beneden & Hesse, 1863 (sensu stricto) (Monogenea: Microcotylidae) and the description of a new species from Dentex dentex (L.) (Teleostei: Sparidae). Parasit Vectors 2020;13:45.
https://doi.org/10.1186/s13071-020-3878-9
- 8. Prosser SW, Velarde-Aguilar MG, León-Règagnon V, Hebert PD. Advancing nematode barcoding: a primer cocktail for the cytochrome c oxidase subunit I gene from vertebrate parasitic nematodes. Mol Ecol Resour 2013;13:1108-1115.
https://doi.org/10.1111/1755-0998.12082
- 9. Larget B, Simon DL, Kadane JB, Sweet D. A Bayesian analysis of metazoan mitochondrial genome arrangements. Mol Biol Evol 2005;22:486-495.
https://doi.org/10.1093/molbev/msi032
- 10. Yoon GH, Shinn A, Sommerville C, Jo JY. Seasonality and the microhabitat of Microcotyle sebastis Goto 1894, a monogenean gill parasite of farmed rockfish, Sebastes schlegeli Hilgendorf 1880. J Aquacult 1998;10:387-394.
- 11. Kim KH, Choi ES, Cho JB, Hwang YJ, Park SI. Influence of temperature on the egg production and hatching of Microcotyle sebastis (Monogenea: Microcotylidae), parasitic on rockfish, Sebastes schlegeli
. J Fish Pathol 1998;11:113-117.
- 12. Kim KH, Park SI, Jee BY. Efficacy of oral administration of praziquantel and mebendazole against Microcotyle sebastis (Monogenean) infestation of cultured rockfish (Sebastes schlegeli). Fish Pathol 1998;33:467-471.
https://doi.org/10.3147/jsfp.33.467
- 13. Park JK, Kim KH, Kang S, Kim W, Eom KS, Littlewood DTJ. A common origin of complex life cycles in parasitic flatworms: evidence from the complete mitochondrial genome of Microcotyle sebastis (Monogenea: Platyhelminthes). BMC Evol Biol 2007;7:11.
https://doi.org/10.1186/1471-2148-7-11
- 14. Scholin CA, Herzog M, Sogin M, Anderson DM. Identification of group-and strain-specific genetic markers for globally distributed Alexandrium (Dinophyceae). Part II. Sequence analysis of a fragment of the LSU rRNA gene. J Phycol 1994;30:999-1011.
https://doi.org/10.1111/j.0022-3646.1994.00999.x
- 15. Nunn GB, Theisen BF, Christensen B, Arctander P. Simplicity-correlated size growth of the nuclear 28S ribosomal RNA D3 expansion segment in the crustacean order Isopoda. J Mol Evol 1996;42:211-223.
https://doi.org/10.1007/BF02198847
- 16. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999;41:95-98.
- 17. Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016;33:1870-1874.
https://doi.org/10.1093/molbev/msw054
- 18. Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003;19:1572-1574.
https://doi.org/10.1093/bioinformatics/btg180
- 19. Page RDM. TREEVIEW: An application to display phylogenetic trees on personal computers. Comput Appl Biosci 1996;12:357-358.
https://doi.org/10.1093/bioinformatics/12.4.357
- 20. Ogawa K. Occurrence of Bivagina tai (Monogenea: Microcotylidae) on the gills of cultured red sea bream Pagrus major
. Bull Japan Soc Sci Fish 1988;54:65-70.
https://doi.org/10.2331/suisan.54.65
- 21. Yamguti S. Systema Helminthum Vol. IV Monogenea and Aspidocotylea. New York. USA Interscience Division, John Wiley & Sons; 1963, p 699.
- 22. Radujković BM, Euzet L. Parasites of marine fishes of Montenegro: monogeneans. Acta Adriat 1989;30:51-135.
- 23. Ono N, Matsumoto R, Nitta M, Kamio Y. Taxonomic revision of Microcotyle caudata Goto, 1894 parasitic on gills of sebastids (Scorpaeniformes: Sebastidae), with a description of Microcotyle kasago n. sp. (Monogenea: Microcotylidae) from off Japan. Syst Parasitol 2020;97:501-516.
https://doi.org/10.1007/s11230-020-09925-5
- 24. Kendall AW Jr. An historical review of Sebastes taxonomy and systematics. Mar Fish Rev 2000;62:1-23.
- 25. Bouguerche C, Gey D, Justine JL, Tazerouti F. Towards the resolution of the Microcotyle erythrini species complex: description of Microcotyle isyebi n. sp. (Monogenea, Microcotylidae) from Boops boops (Teleostei, Sparidae) off the Algerian coast. Parasitol Res 2019;118:1417-1428.
https://doi.org/10.1007/s00436-019-06293-y
- 26. Bucklin A, Steinke D, Blanco-Bercial L. DNA barcoding of marine metazoa. Annu Rev Mar Sci 2011;3:471-508.
https://doi.org/10.1146/annurev-marine-120308-080950