Cloning of the novel putative apoptosis-related gene of Spirometra erinacei (Order Pseudophyllidea)
Article information
Abstract
We postulated that apolysis was processed in accordance with apoptotic changes occurring in a cestode, Spirometra erinacei (Pseudophyllidea). We cloned the novel putative apoptosis-associated gene from S. erinacei via screening of a S. erinacei cDNA library with a ced-3 gene (activator of apoptosis) probe from Caenorhabditis elegans. We identified a 261-bp cDNA sequence, which encodes for an 86-amino acid protein. The cloned gene expression was observed in the neck and gravid proglottids via Northern blotting, using cloned cDNA inserts as probes, but the clone was not expressed in any of other tissues. We suggest that this gene may be involved in the apolysis of S. erinacei during normal tissue development and differentiation in cestode parasites.
Spirometra erinacei inhabits the intestines of cats and dogs. Apolysis, or the expulsion of gravid proglottids, occurs during developmental periods, and functions to accelerate the dissemination of eggs. Apoptosis, or programmed cell death (PCD), plays a key role in developmental biology and maintenance of the steady state in continuously renewing tissues, particularly in embryogenesis, normal tissue development, organ involution, and differentiation (Ameisen et al., 1995). Apoptosis is a specific mode of cell death, which can be identified by virtue of a characteristic pattern of morphological, biochemical, and molecular changes (Kerr et al., 1972).
The ced-3 gene functions in a cell-autonomous manner, inducing PCD in C. elegans, and encodes for a protein which is a member of the interleukin-1β (IL-1β-converting enzyme (ICE, caspase-1 family) cysteine protease family (Yuan and Horvitz; 1990; Thornberry et al., 1992; Yuan et al., 1993). The cloned ced-3 gene of C. elegans has been demonstrated to encode for proteins exhibiting a great deal of similarity to vertebrate cell death genes, and analysis of this gene has demonstrated that both nematodes and mammals share a common pathway for PCD (Hengartner and Horvitz, 1994). We postulated that apolysis might occur as the result of an apoptotic change in cestodes, for example, S. erinacei. Apolysis is an important phenomenon, as it is the method by which cestodes reproduce. The primary objective of this study was to clone the apoptosis-related gene of S. erinacei. Thus, we cloned the apoptosis-related gene of S. erinacei, using a ced-3 gene probe from C. elegans.
Adult worms of S. erinacei were obtained from the intestines of dogs at 4 weeks post-infection with spargana from snakes (Rhadophis tigrinus tigrinus, Boie, 1826) in Korea. The adult specimens were stored at -70℃ until required. We employed a cDNA synthesis kit (Stratagene, La Jolla, California, USA) for cDNA synthesis and for construction of the S. erinacei cDNA library. The total RNA was isolated using the RNAgents total RNA isolation system (Promega, Madison, Wisconsin, USA) and mRNA was isolated using the PolyATtract® mRNA isolation system I (Promega). The procedure in all its details was executed in accordance with the manufacturer's recommendations.
Previously extracted mRNA (5 µg) and first-strand methyl nucleotide mixture and linker-primer (1.4 µg/µl) were employed in the synthesis of first-strand cDNA, and the reaction was catalyzed using MMLV (molony murine leukemia virus) reverse transcriptase for 1 hr at 37℃. Second-strand synthesis was accomplished using the above first-strand mixture along with 10 x second-strand buffer, second-strand dNTP mixture, [α·-32P] dATP (800 Ci/mmol), RNase H (1.5 U/µl), and DNA polymerase I (9 U/µl) for 2.5 hr at 16℃. The cDNA terminal was blunted with blunting dNTP mix, after which it was cloned using Pfu DNA polymerase (2.5 U/µl) at 72℃ for a 30-min reaction, then extracted using phenol-chloroform and precipitated with ethanol. An EcoR1 adaptor was ligated using blunted cDNA and T4 DNA ligase. The blunted cDNA (100 ng) was then ligated into the Uni-ZAP XR vector (1-2 µg) arms, and the ligation mixture was packaged using the Gigapack III gold Packaging System (Stratagene). Appropriate dilutions of the packaging extracts and the host E. coli were mixed, poured onto NZY agar plates, and incubated for 6-8 hr at 37℃, after which the numbers of plaques were counted. This procedure, in detail, was included in the manufacturer's cDNA library synthesis kit (Stratagene).
The ced-3 cDNA plasmid probe (cloned into pBluescript SK (-) vector, insert size: 2,482 bp) of C. elegans was obtained from Dr. H. Robert Horvitz (Howard Hughes Medical Institute, Department of Biology and McGovern Institute for Brain Research, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA) (Yuan et al., 1993). The plasmid was digested with BamHI and the purified extracted DNA fragments were labeled using an enhanced chemiluminescent (ECL) direct nucleic acid labeling system (Amersham, Buckinghamshire, UK) in accordance with the manufacturer's instructions. 50,000 plaques of the amplified S. erinacei cDNA library were screened using the ECL-labeled ced-3 cDNA probes. Both prehybridization and hybridization were conducted at 42℃, as previously described by Sambrook and Russell (2001).
Several clones confirmed to positively react with the probe were then isolated and subjected to further screening in order to verify positivity. The presence of the insert DNA (approximately 550 bp, data not shown) was confirmed via PCR using two universal primers. The positive clones were isolated using an in vivo excision protocol, using the ExAssit helper phage with the SOLA strain, in accordance with the instructions provided by the manufacturers (Stratagene). In order to confirm the cDNA insert fragment size, PCR was conducted using two universal primers (T7 and T3, each at 10 pmole), Taq DNA polymerase (Takara Bio, Otsu, Shiga, Japan), and positive DNA clones encoding for the putative apoptosis gene of S. erinacei. PCR amplification was conducted in a DNA thermal cycler (Applied Biosystem GeneAmp PCR System, Foster, California, USA) over 30 cycles (94℃ for 40 sec, 55℃ for 30 sec, and 72℃ for 1 min 30 sec), with a 6-minute final extension step at 72℃. DNA inserts from the phagemids were purified with a plasmid miniprep kit (Qiagen, Valencia, California, USA). DNA sequencing was conducted via dideoxy chain termination (Sanger et al., 1977). The obtained sequence analysis was then compared with the nucleotide and amino acid sequence data in the GenBank and BLAST databases of the National Center for Biotechnology Information (NCBI).
In order to compare the identity of the obtained sequence with those of other reportedly apoptosis-associated genes, we obtained other genes via a NCBI GenBank search, and also conducted multiple sequence alignments using the Clustal W program from the European Bioinformatics Institute (EBI) (http://www.ebi.ac.uk/clustalw) (Higgins et al., 1994). We predicted the presence and location of signal peptide cleavage sites on the basis of a combination of several artificial neural networks, using the SignalP 3.0 Server program at EBI (http://www.cbs.dtu.dk/services/SignalP/) (Bendtsen et al., 2004). The phosphorylation site and N-myristoylation site were predicted using the PROSITE protein families and domains database (http://au.expasy.org/prosite/) (Hulo et al., 2004). For Northern blotting, total RNA and mRNA from S. erinacei adult tissues (neck, immature, mature and gravid proglottids) were extracted using the same commercial kit as above (Promega). Total RNA (15 µg) were separated on 1.0% formaldehyde agarose gel, and transferred to membranes, then hybridized as described by Sambrook and Russel (2001). The cloned cDNA insert (approximately 550 bp) via PCR of S. erinacei, which was used as a probe (25 to 100 ng), was randomly-radiolabeled with [α·-32P] dCTP using a T7 QuickPrime Labeling Kit (Amersham Pharmacia Biotech Co., Denver, Colorado, USA), then employed as a probe for hybridization of the RNA onto the membrane.
Approximately 50,000 clones were screened in total, and 40 were determined to react with the C. elegans ced-3 cDNA probe. These plaques were then purified and re-tested, and 2 of them remained positive. In order to confirm the positive clones by PCR, pBluescript phagemids from the 2 clones were isolated via in vivo excision, and the plasmid was purified with a mini-prep kit, after which we conducted PCR using universal primer sets. This revealed that the 2 clones harbored inserts of approximately 550 bp (data not shown). Partial sequencing from the cloned positive clones revealed that the sequences of the 2 clones were identical. The characterization of completed sequences of the 2 clones revealed that they harbored inserts of 408 bp. This sequence consisted of a 36-bp 5' untranslated region, a 261-bp putative open reading frame (ORF), and 110-bp of a 3'-untranslated region, which included a putative polyadenylation signal (AATAAA) and a poly-A tail. The longest ORF encoded for an 86-amino acid polypeptide. The putative signal peptide cleavage site, phosphorylation site, and N-myristoylation site involved in apoptosis and proteolytic processing are shown in Fig. 1. Apoptosis-related gene expression was observed in the neck and gravid proglottids via Northern blotting using the cloned cDNA insert as a probe, but this gene was not expressed in any other tissues (Fig. 2).

Nucleotide sequence and deduced amino acid sequence of the cloned cDNA from Spirometra erinacei in the pBluescript SK (-) phagemid. The total insert sequence was 408 bp (-36 to 372) in length and consisted of 36 bp (-36 to -1) of an 5' untranslated region, 261 bp (1 to 261) of a putative open reading frame (ORF), and 110 bp (262 to 372) of a 3'-untranslated region, including a putative polyadenylation signal (underlined AATAAA, 321 to 326) and a poly-A tail (underlined 28 bp, 345 to 372). The longest ORF encodes for an 86-amino acid polypeptide. The numbers at both sides indicate nucleotide positions (left side) and amino acid positions (right side). The start and stop codons are indicated by asterisks (*). EcoRI and XhoI indicate the cloning sites at both ends (bold italics). The predicted signal peptide cleavage sites in the amino acid sequence are boxed in grey at positions 15-16 (AL) and 20-21 (AS). The predicted phosphorylation sites are boxed in gray at position 30-33 (TDEE). A predicted N-myristoylation site is boxed in gray at position 66-71 (GLIEGA).
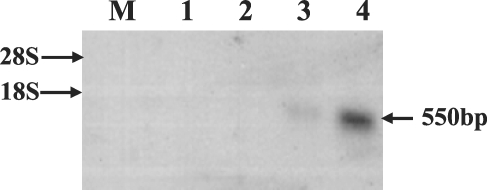
Northern blot of S. erinacei total RNA using the putative cloned cDNA as a probe. Lane M: marker (18S +28S ribosomal RNA, 8 µg, Sigma), lane 1: immature proglottid (15 µg), lane 2: mature proglottid (15 µg), lane 3: neck (15 µg), and lane 4: gravid proglottid (15 µg). Arrow size indicates approximately 550 bp.
Apoptosis, or PCD, is an essential process during the normal development and homeostasis of all multicellular organisms. Recently, several human and murine ICE or caspase homologies have been cloned (Yuan and Horvitz; 1990). The ced-3 gene of C. elegans encodes for a cysteine protease that is essential for developmentally-regulated apoptosis. Oligonucleosomal DNA fragmentation, a characteristic feature of the apoptotic program from genomic DNA, was also observed in the whole adult part of S. erinacei (data not shown), and the cloned gene was expressed weakly in the neck and abundantly expressed in the gravid proglottids of S. erinacei tissues, according to the results of Northern blotting (Fig. 2). Expressions in gravid proglottids and neck may be related to cycle of the degeneration and regeneration of S. erinacei, i.e. natural expulsion of gravid proglottids as well as expulsion of entire proglottids followed by regeneration of proglottids in the host such as the dog.
Using our obtained data, we predicted the presence and location of putative signal peptide cleavage sites in amino acid sequences, using the SignalP 3.0 Server, with the help of published papers regarding similar data on prokaryotic and eukaryotic organisms (Nielsen et al., 1997). SignalP, currently one of the most extensively employed methods, predicts the presence of signal peptidase I cleavage sites, on the basis of a combination of several artificial neural networks (NN) and hidden Markov models (HMM) (Bendtsen et al., 2004). We detected a positively charged n-region (H at position 14), a hydrophobic region (AL at position 15, 16; A at position 20), and a neural but polar region (S at position 21). This predictive performance is relevant to interpretations of experimental data, which are not always perfect. We also detected putative potential myristoylated glycine residues within the C-terminal residue, at position 66-71 (GLIEGA). We have also surmised that myristate may play a role in mediation of protein-protein interactions, as the majority of eukaryotic proteins are acylated via the covalent addition of myristate (a C14-saturated fatty acid) to their N-terminal residues, via an amide linkage. The acylation of the enzyme is responsible for both post-translational modification and localization (Grand, 1989). This gene may be involved in apolysis, as apoptotic-related gene expression was detected in S. erinacei.
We were unable to detect any conserved pentapeptide active sites (GACRG positions 356-360 of the CED-3 protein, Craen et al., 1997) in the putative cloned cDNA, as our partial nucleotide sequence evidenced a high degree of homology (46%, 119/261 bp) with the ced-3 gene of C. elegans within the N-terminal regions (data not shown). The discrepancy between the apparent 550 bp yielded by PCR and Northern blotting and the final 408 bp insert can be explained by the fact that T3 and T7 primers exist outside the insert within the pBluescript phagemid. This experiment with cestodes permitted us to evaluate the role of cysteine protease in apoptosis. This information may also provide us with a great deal of insight into the evolutionary role of regulated death programming in the context of natural selection.
References
Notes
This study was supported by the Grant-in-Aid from Korea Science and Engineering Foundation, 1996-1999 (Grant No. 96-04-03-05-01-3)
Nucleotide sequences data reported in this paper are available in GenBankTM database under the accession number AF229035.