Abstract
Drug resistance is an important problem hindering malaria elimination in tropical areas. Point mutations in Plasmodium falciparum dihydrofolate reductase (Pfdhfr) and dihydropteroate synthase (Pfdhps) genes confer resistance to antifolate drug, sulfadoxine-pyrimethamine (SP) while P. falciparum chloroquine-resistant transporter (Pfcrt) genes caused resistance to chloroquine (CQ). Decline in Pfdhfr/Pfdhps and Pfcrt mutations after withdrawal of SP and CQ has been reported. The aim of present study was to investigate the prevalence of Pfdhfr, Pfdhps, and Pfcrt mutation from 2 endemic areas of Thailand. All of 200 blood samples collected from western area (Thai-Myanmar) and southern area (Thai-Malaysian) contained multiple mutations in Pfdhfr and Pfdhps genes. The most prevalent haplotypes for Pfdhfr and Pfdhps were quadruple and double mutations, respectively. The quadruple and triple mutations of Pfdhfr and Pfdhps were common in western samples, whereas low frequency of triple and double mutations was found in southern samples, respectively. The Pfcrt 76T mutation was present in all samples examined. Malaria isolated from 2 different endemic regions of Thailand had high mutation rates in the Pfdhfr, Pfdhps, and Pfcrt genes. These findings highlighted the fixation of mutant alleles causing resistance of SP and CQ in this area. It is necessary to monitor the re-emergence of SP and CQ sensitive parasites in this area.
-
Key words: Plasmodium falciparum, dihydropteroate synthase, dihydrofolate reductase, chloroquine-resistant transporter, molecular marker
INTRODUCTION
Although malaria is an ancient disease caused by
Plasmodium parasite, it remains important to public health to present era.
Plasmodium falciparum infection causes variable clinical symptoms ranging from asymptomatic to severe manifestations. The emergence of resistance of
P. falciparum to the available antimalarial drugs is an important factor for malaria control [
1]. In Thailand, resistance to many antimalarial drugs, including chloroquine (CQ), sulfadoxine-pyrimethamine (SP), mefloquine, and artemisinin has been reported [
2,
3]. CQ resistant
P. falciparum was reported in the early 1960s [
4,
5]. In 1973, SP replaced CQ as the first-line treatment for uncomplicated falciparum malaria due to widespread resistance [
1,
6], but after 10 years, SP was ineffective [
1,
7]. Then, mefloquine was introduced in 1985 and resistance emerged in the same decade [
8]. Artemisinin-based combination therapy was introduced as first-line treatment in 1995 [
9].
Molecular epidemiological investigation provides information for detecting the emergence and spread of antimalarial drug resistance. Mutations in the
P. falciparum dihydrofolate reductase (
Pfdhfr) and
P. falciparum dihydropteroate synthase (
Pfdhps) genes (at codons 51, 59, 108, and 164 of
Pfdhfr and 437, 540, and 581 of
Pfdhps) are associated with SP treatment failures [
10,
11]. The mutations in
Pfdhfr and
Pfdhps genes were staged, resulting in increased levels of SP drug resistance [
12]. The
P. falciparum CQ resistance transporter gene (
Pfcrt) K76T mutation has been linked to
P. falciparum CQ resistance [
13].
In some countries, the withdrawal of CQ for
P. falciparum treatment,
Pfcrt mutation (K76T) gently decreased and disappeared completely [
14–
17]. Similar to withdrawal of SP for
P. falciparum treatment,
Pfdhfr and
Pfdhps gene mutations also decreased in some countries [
18–
21]. Conversely, alleles conferring CQ and SP resistance still occur at high frequency after discontinuation of these drugs [
22,
23]. However, declining of
Pfdhfr,
Pfdhps, and
Pfcrt mutations might be associated with duration of drug withdrawal and geographical differences.
The objective of the present study was to investigate the prevalence of 5 Pfdhfr (A16V, N51I, C59R, S108N/T, and I164L), 5 Pfdhps (S436A, A437G, K540E, A581G, and A613S/T) and 1 Pfcrt (K76T) mutation from 2 different endemic areas of Thailand.
MATERIALS AND METHODS
Ethics approval
The study protocol was reviewed and approved by the Ethics Committee of Thammasat University (COA No. 134/2561). All patients were informed about the study objectives, sampling technique, and the benefits of the study. Informed consents were obtained according to the ethical standards from all patients.
Sample collection
A total of 200 dried blood spot samples were collected during 2007–2017 from patients with P. falciparum infection who attended malaria clinics in the western (Tak Province) and southern (Yala Province) regions along the Thai-Myanmar and Thai-Malaysian border, respectively.
Extraction of parasite genomic DNA
Genomic DNA of all blood samples was prepared using a QIAamp DNA extraction mini-kit (QIAGEN, Valencia, California, USA) according to manufacturer’s instruction and used as a template for polymerase chain reaction (PCR) amplification.
Amplification and detection of the Pfdhfr and Pfdhps
Pfdhfr and
Pfdhps genes were amplified by nested PCR using
Pfdhfr and
Pfdhps specific primers (
Table 1) according to the previously described methods with some modification [
24]. Briefly, the PCR was carried out with the following reaction mixture including 0.25 μM of each primer, 1.5 mM MgCl
2 (Thermo scientific, Waltham, Massachusetts, USA), 1×Taq buffered with KCl (Thermo scientific), 200 μM deoxynucleotides (dNTPs) (Bioline, London, UK), 2 μl of genomic DNA in the primary PCR, and 1 μl of primary PCR product in nested PCR and 1 unit of Taq DNA polymerase (Thermo scientific). All of the PCR products were then analyzed on 1% agarose gel and visualized under UV illuminator. PCR products were digested with restriction enzymes (
Table 1) [
24] then the restriction fragments were analyzed on 1.2% agarose gel and visualized under UV illuminator.
Amplification and detection of the Pfcrt
Amplification of K76T was performed by nested PCR using
Pfcrt specific primers (
Table 1) according to the previously described methods with some modification [
25–
27]. Briefly, the PCR was carried out with the following reaction mixture including 0.1 μM of each primer, 2.5 mM MgCl
2 (Thermo scientific), 1×Taq buffered with KCl (Thermo scientific), 100 μM deoxynucleotides (dNTPs) (Bioline), 0.5 μl of genomic DNA in the primary PCR, and 0.5 μl of primary PCR product in nested PCR and 0.5 unit of Taq DNA polymerase (Thermo scientific). All of the PCR products were then analyzed on 1.5% agarose gel and visualized under UV illuminator. PCR products were digested with restriction enzymes ApoI (New England Biolabs Inc., Hertfordshire, UK) (
Table 1), as described by the manufacturer. Then the restriction fragments were analyzed on 2.0% agarose gel and visualized under UV illuminator.
Data analysis was performed by SPSS software version 21.0 (IBM Corporation, Armonk, New York, USA). The chi-square test was used to compare the frequencies and correlations of all data. The level of significance was set at P<0.05.
RESULTS
Analysis of Pfdhfr and Pfdhps mutations
A total of 200
P. falciparum samples were successfully amplified and analyzed for both the
Pfdhfr and
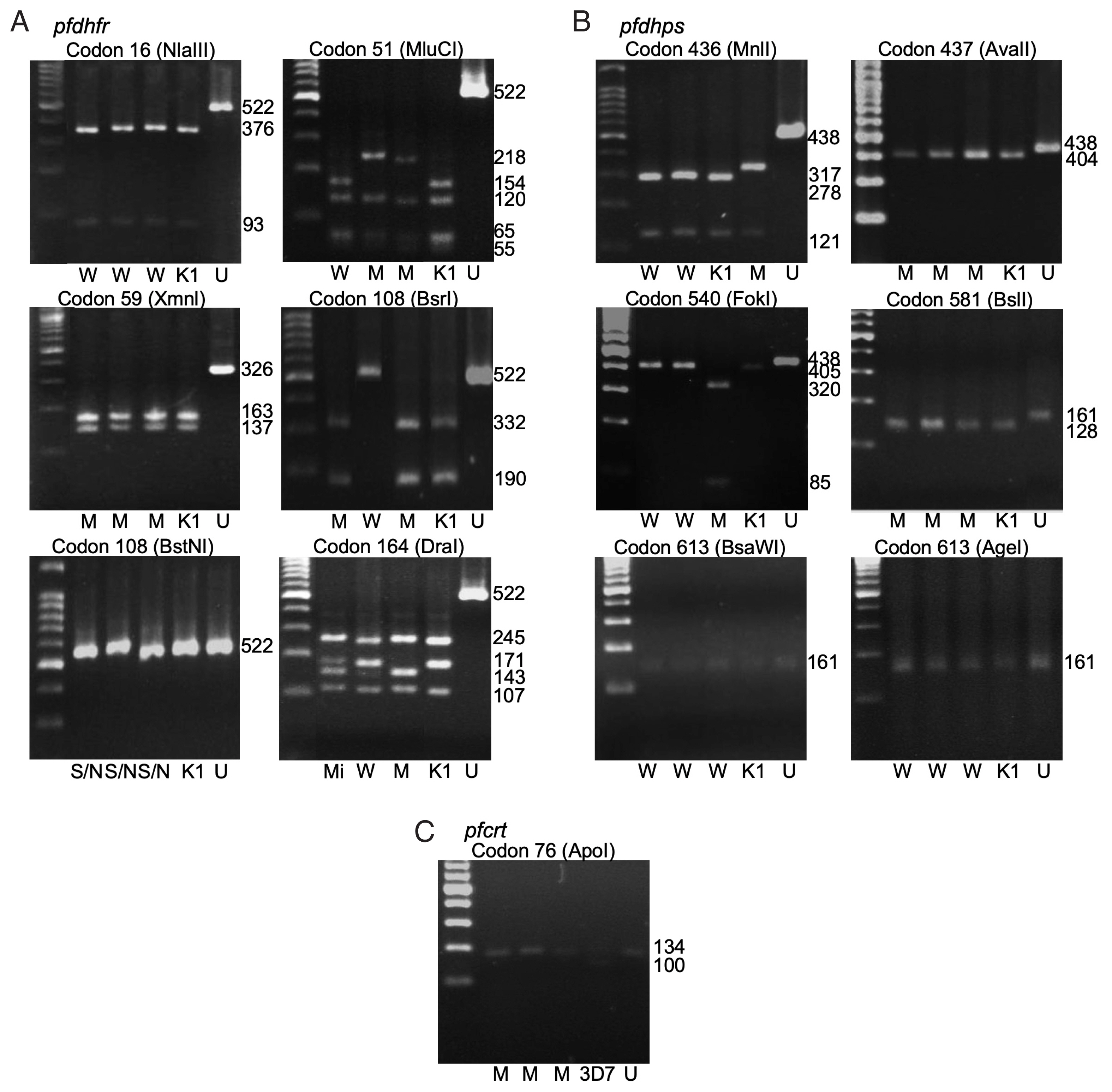
Pfdhps genes. The polymorphisms at each codon were demonstrated by restriction fragments (
Fig. 1A, B). The frequencies of
Pfdhfr and
Pfdhps mutations was summarised in
Table 2. All samples had at least 1 codon mutation in the
Pfdhfr (A16V, N51I, C59R, S108N/T, and I164L) and
Pfdhps (S436A, A437G, K540E, A581G, and A613S/T). Four codon mutations were detected in
Pfdhfr (51I, 59R, 108N, and 164L) in samples from 2 areas. All isolates carried mutations at codon 59 and 108 in
Pfdhfr. Four codon mutations were detected in
Pfdhps (436A, 437G, 540E, and 581G) in samples from Tak Province, whereas 3 codon mutations were detected from Yala Province. All isolates carried wild-type alleles at codon 613 in
Pfdhps. Mixed genotypes were detected in thirty isolates by codon 51 and 164 of
Pfdhfr, and 436 and 540 of
Pfdhps that were no processed further. There was no wildtype allele ANCSI, but 3 alleles (AIRNI, AIRNL, and ANRNL) of
Pfdhfr were identified in this study (
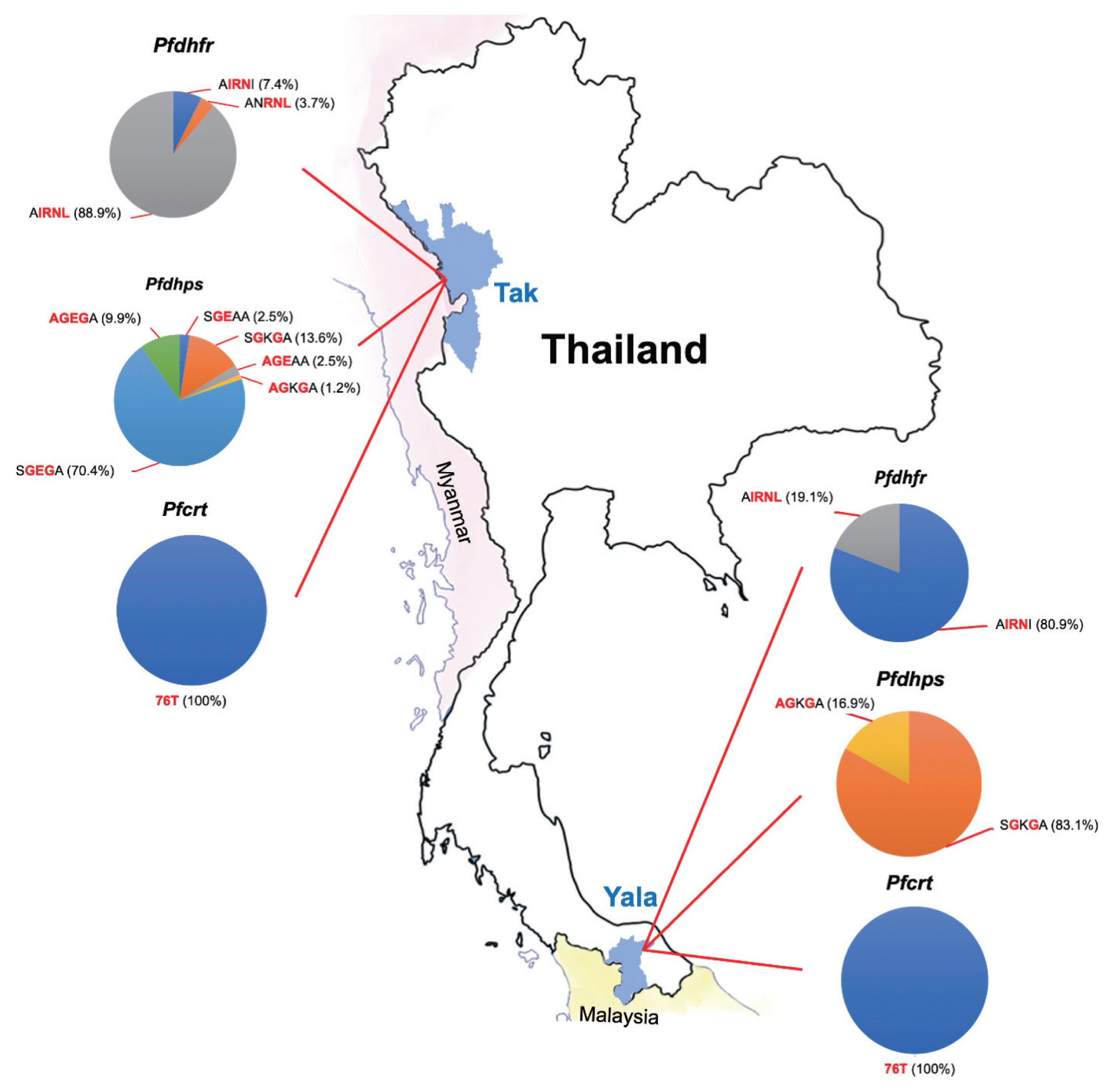
Table 3). AIRNL was the most prevalent allele (52.4%) in all
P. falciparum isolates collected from Tak Province (88.9%) (
Fig. 2). AIRNI was the most prevalent allele in
P. falciparum isolates from Yala Province (80.9%). Statistical significance was found between 2 study areas (
P<0.001). For
Pfdhps, 6 alleles were identified (
Table 3). SGKGA was the most prevalent allele (50.0%) found in
P. falciparum isolates from Yala Province (83.1%). SGEGA was the most prevalent allele in
P. falciparum isolates from Tak Province (70.4%). Statistical significance was found between 2 study areas (
P<0.001). Eleven allele combination of
Pfdhfr-
Pfdhps were found in this study (
Table 4). The quintuple mutation (AIRNI-SGKGA), which comprise triple mutations in
Pfdhfr and 2 mutations in
Pfdhps were found to be most prevalent in study population (35.3%) and in isolates from Yala Province (66.3%). The septuple mutation (AIRNL-SGEGA), which comprised quadruple mutations in
Pfdhfr and 3 mutations in
Pfdhps were most frequent in isolates from Tak Province (64.2%).
A total of 187 samples (93.5%) were analyzed by nested PCR for the
Pfcrt K76T gene. The
Pfcrt mutation resulting in substitution of threonine (T) for lysine (K) at position 76 was present in all studied samples from 2 endemic areas (
Figs. 1C,
2).
DISCUSSION
Mutations on
Pfdhfr and
Pfdhps genes associated with SP resistance have been reported in several malaria endemic areas such as Guinea [
28], Indonesia [
29], Malaysia [
30], Myanmar [
31], and Thailand [
32,
33]. In Thailand, a previous study revealed that the change of
Pfdhfr point mutations from double mutations to triple and quadruple mutations in some areas [
34,
35] which the number of mutations is correlated with increased level of SP resistance [
12]. In the present study, all
P. falciparum isolates had at least three mutation point in
Pfdhfr genes, indicated persistence of highly mutations on SP resistant markers. The
Pfdhps mutation studies conducted between 2001 and 2007 in Thailand indicated that there has been fluctuation of the
Pfdhps mutations between triple and quadruple mutations. In this study, predominant frequency of double mutation was found especially in isolates from southern endemic area. The predominance of triple mutations was also found in isolates from western area. This result indicated that high mutation in
Pfdhfr and
Pfdhps genes with different frequency existed in these 2 different localities. In Thailand, SP was withdrawn from
P. falciparum treatment for many years. Although decreased of
Pfdhfr and
Pfdhps mutations were reported from some countries after the withdrawal of these drugs, high frequency of
Pfdhfr and
Pfdhps mutations still present in Thailand. This existence of mutations on
Pfdhfr and
Pfdhps genes may be associated with using of other antifolate drugs that can also induce pressure on
Pfdhfr and
Pfdhps.
A previous study has demonstrated that high prevalence of
Pfcrt K76T mutation in study isolates might contribute to CQ resistance to
P. falciparum [
36]. A high prevalence rate of
Pfcrt K76T mutation was previously observed in several countries such as Mali [
25], Kenya [
37], Indonesia [
26], Philippines [
38] and Thailand [
39–
41]. Declining of
Pfcrt mutations after withdrawal of CQ has been reported in Malawi [
14], Tanzania [
15], Kenya [
16], and China [
17]. However, even CQ was withdrawn from Thailand for long period, the CQ resistance allele still remains with high frequency. This complete fixation of CQ resistance in
P. falciparum is might due to the co-existence of
P. falciparum and
P. vivax infections in this country while CQ is a standard regimen for
P. vivax malaria treatment, leading to the phenomenon of continuous exposure to drug pressure in
P. falciparum.
Our study demonstrates a high prevalence of Pfdhfr, Pfdhps, and Pfcrt mutations of P. falciparum isolates from 2 endemic areas in Thailand, emphasizing the fixation of mutant Pfdhfr, Pfdhps and Pfcrt alleles that confer consistent resistance of SP and CQ. SP and CQ drugs are still not appropriate for P. falciparum treatment in Thailand and other antimalarial groups should be considered.
Notes
-
The authors have no conflict of interest.
ACKNOWLEDGMENT
This research was supported by the Program Management Unit for Human Resources and Institutional Development, Research and Innovation, NXPO [grant number B05F630043].
Fig. 1The polymorphism of pfdhfr (A), pfdhps (B), and pfcrt (C) gene by gel electrophoresis. W-wildtype, M-mutant, S/N-serine/threonine, Mi-mixed, K1-P. falciparum K1 strain, 3D7- P. falciparum 3D7 strain, U-undigested fragment. Fragment sizes in base pair (bp) are shown.
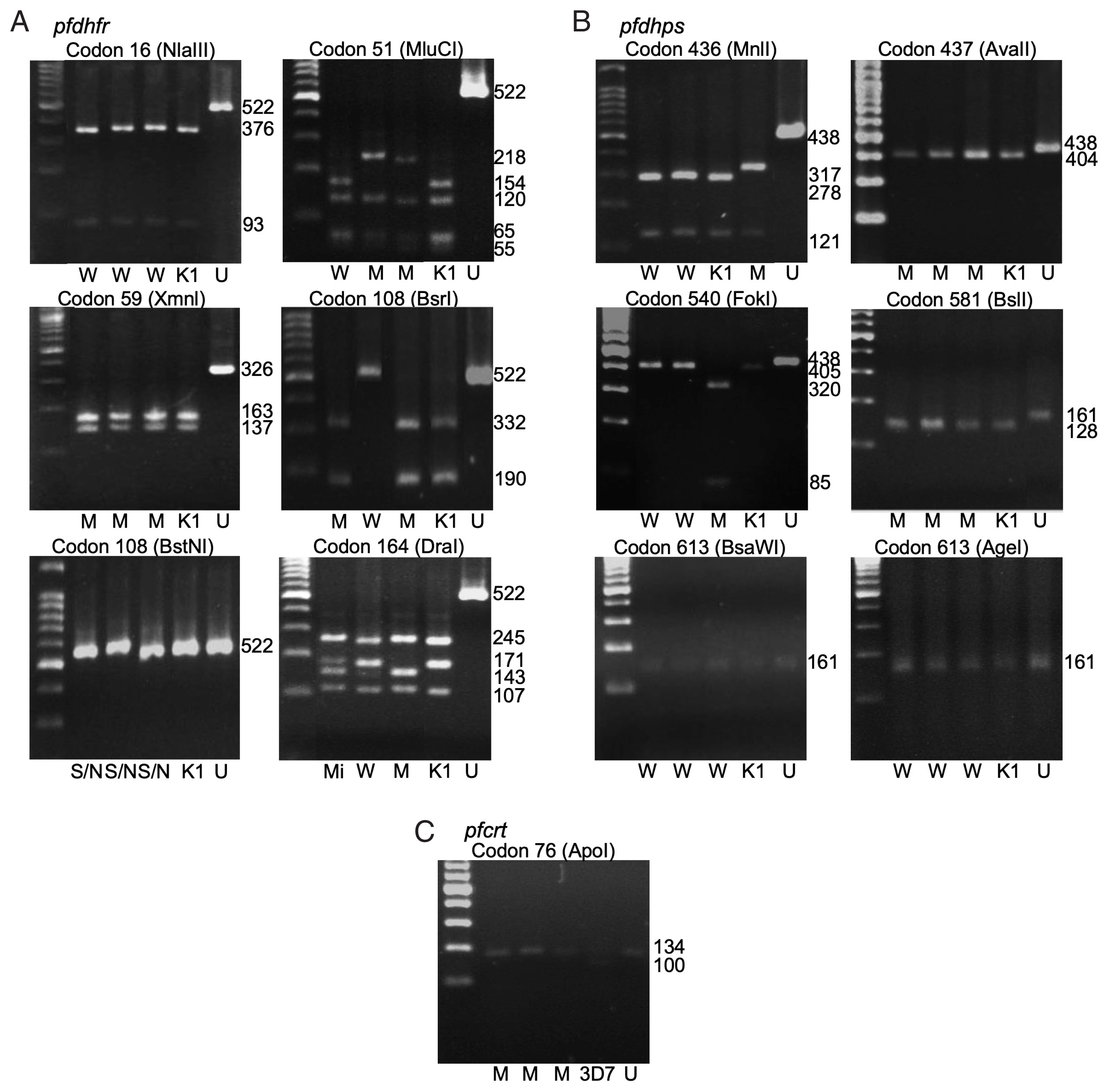
Fig. 2The proportions of mutations in 3 resistance genes (pfdhfr, pfdhps, and pfcrt) observed in P. falciparum isolates in this study.
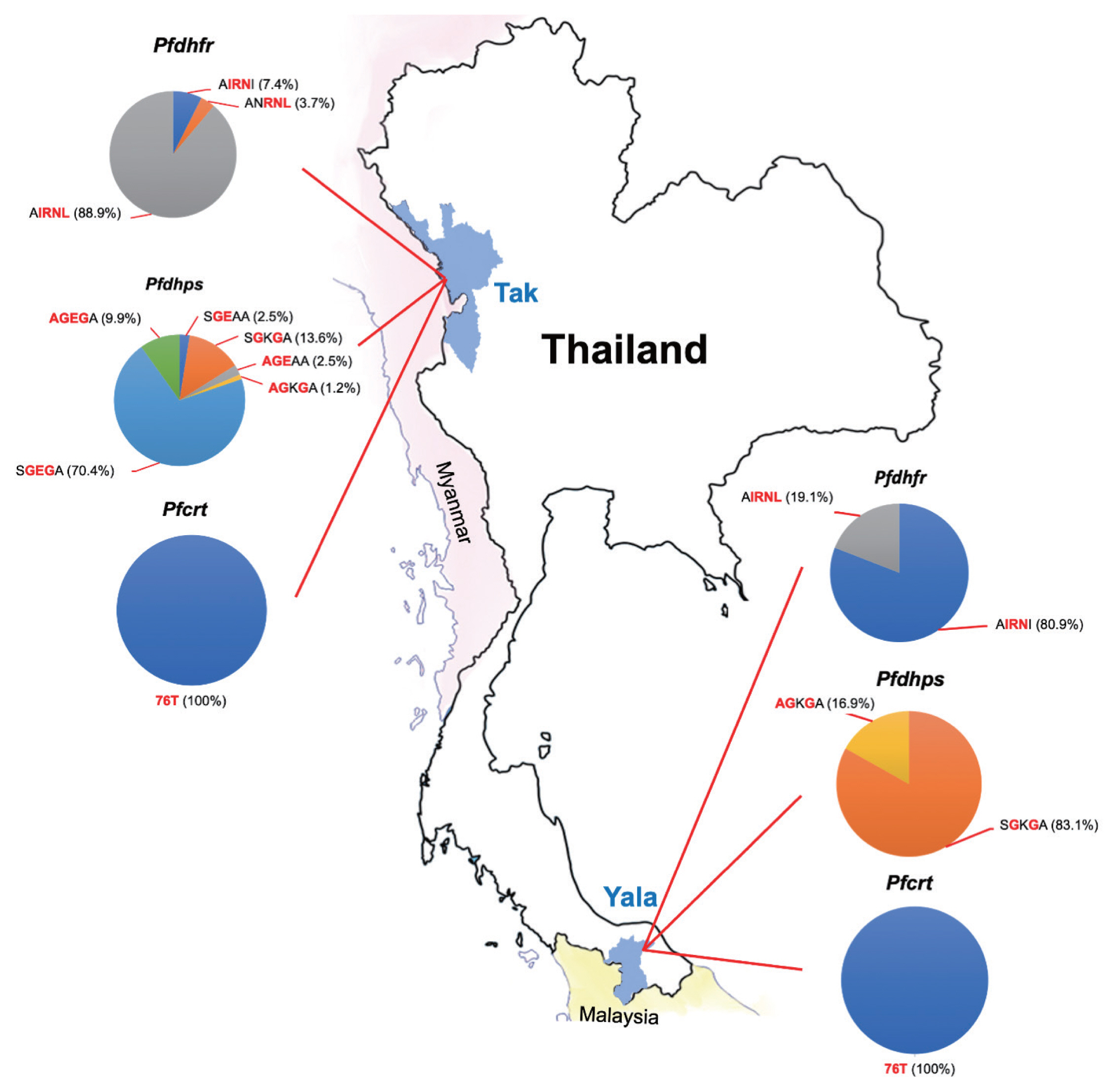
Table 1The primers and enzymes for genotyping of Pfdhfr, Pfdhps and Pfcrt genes
Table 1
|
Gene |
PCR |
Primer |
Primer sequence (5′ to 3′) |
RFLP position |
Restrictionenzyme |
PCR size (bp) |
Restriction product size (bp) |
|
|
Wild type |
Mutation |
|
Pfdhfr
|
Primary |
M1 |
TTTATGATGGAACAAGTCTGC |
|
|
|
|
|
|
|
M5 |
AGTATATACATCGCTAACAGA |
|
|
|
|
|
|
Secondary |
M3 |
TTTATGATGGAACAAGTCTGCGACGTT |
A16V |
Nlalll |
522 |
376, 93, 53 |
376, 146 |
|
(16, 51, 108, 164) |
F/ |
AAATTCTTGATAAACAACGGAACCTTTTA |
N51I |
MluCI |
|
154, 120, 65, 55 |
218, 120, 65, 55 |
|
|
|
|
S108T |
BstNI |
|
522 |
181, 145 |
|
|
|
|
S108N |
Bsrl |
|
522 |
332, 190 |
|
|
|
|
I164L |
DraI |
|
245, 171, 107 |
245, 143, 107, 27 |
|
Secondary |
F |
GAAATGTAATTCCCTAGATATGGAATATT |
C59R |
Xmnl |
326 |
189, 137 |
163, 137, 26 |
|
(59) |
M4 |
TTAATTTCCCAAGTAAAACTATTAGAGCTTC |
|
|
|
|
|
|
|
Pfdhps
|
Primary |
R2 |
AACCTAAACGTGCTGTTCAA |
|
|
|
|
|
|
|
R/ |
AATTGTGTGATTTGTCCACAA |
|
|
|
|
|
|
Secondary |
K |
TGCTAGTGTTATAGATATAGGATGAGCATC |
S436A |
MnlI |
438 |
317, 121 |
278, 121, 39 |
|
(436, 437, 540) |
K/ |
CTATAACGAGGTATTGCATTTAATGCAAGAA |
A437G |
AvaII |
|
438 |
404, 34 |
|
|
|
|
K540E |
FokI |
|
405, 33 |
320, 85, 33 |
|
Secondary |
L |
ATAGGATACTATTTGATATTGGACCAGGATTCG |
A581G |
BslI |
161 |
161 |
128, 33 |
|
(581, 613) |
L/ |
TATTACAACATTTTGATCATTCGCGCAACCGG |
A613S |
BsaWI |
|
161 |
131, 30 |
|
|
|
|
A613T |
AgeI |
|
161 |
128, 33 |
|
|
Pfcrt
|
Primary |
CRTP1 |
CCGTTAATAATAAATACACGCAG |
|
|
|
|
|
|
|
CRTP2 |
CGGATGTTACAAAACTATAGTTACC |
|
|
|
|
|
|
Secondary |
CRTD1 |
TGTGCTCATGTGTTTAAACTT |
K76T |
ApoI |
134 |
100, 34 |
134 |
|
|
CRTD2 |
CAAAACTATAGTTACCAATTTTG |
|
|
|
|
|
Table 2Prevalence of Plasmodium falciparum dihydrofolate reductase (Pfdhfr) and dihydropteroate synthase (Pfdhps) single nucleotide polymorphisms (SNPs) in 200 P. falciparum isolates from 2 endemic areas of Thailand
Table 2
|
Gene |
Amino acid position |
SNPs |
Prevalence (%) |
P-value |
|
|
Total n=200 |
Tak Province n=100 |
Yala Province n=100 |
|
Pfdhfr
|
16 |
A (wild-type) |
200 (100.0) |
100 (100.0) |
100 (100.0) |
- |
|
|
V (mutant) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
|
|
51 |
N (wild-type) |
3 (1.5) |
3 (3.0) |
0 (0.0) |
0.001a
|
|
|
I (mutant) |
187 (93.5) |
87 (87.0) |
100 (100.0) |
|
|
|
M (mix) |
10 (5.0) |
10 (10.0) |
0 (0.0) |
|
|
59 |
C (wild-type) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
- |
|
|
R (mutant) |
200 (100.0) |
100 (100.0) |
100 (100.0) |
|
|
108 |
S (wild-type) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
- |
|
|
T (mutant) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
|
|
|
N (mutant) |
200 (100.0) |
100 (100.0) |
100 (100.0) |
|
|
164 |
I (wild-type) |
84 (42.0) |
6 (6.0) |
78 (78.0) |
<0.001a
|
|
|
L (mutant) |
103 (51.5) |
86 (86.0) |
17 (17.0) |
|
|
|
M (mix) |
13 (6.5) |
8 (8.0) |
5 (5.0) |
|
|
|
Pfdhps
|
436 |
S (wild-type) |
158 (79.0) |
79 (79.0) |
79 (79.0) |
0.946 |
|
|
A (mutant) |
29 (14.5) |
14 (14.0) |
15 (15.0) |
|
|
|
M (mix) |
13 (6.5) |
7 (7.0) |
6 (6.0) |
|
|
437 |
A (wild-type) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
- |
|
|
G (mutant) |
200 (100.0) |
100 (100.0) |
100 (100.0) |
|
|
540 |
K (wild-type) |
116 (58.0) |
16 (16.0) |
100 (100.0) |
<0.001a
|
|
|
E (mutant) |
83 (41.5) |
83 (83.0) |
0 (0.0) |
|
|
|
M (mix) |
1 (0.5) |
1 (1.0) |
0 (0.0) |
|
|
581 |
A (wild-type) |
7 (3.5) |
7 (7.0) |
0 (0.0) |
0.007a
|
|
|
G (mutant) |
193 (96.5) |
93 (93.0) |
100 (100.0) |
|
|
613 |
A (wild-type) |
100 (100.0) |
100 (100.0) |
100 (100.0) |
- |
|
|
S/T (mutant) |
0 (0.0) |
0 (0.0) |
0 (0.0) |
|
Table 3
Plasmodium falciparum dihydrofolate reductase (Pfdhfr) and dihydropteroate synthase (Pfdhps) alleles in 170 P. falciparum isolates from 2 endemic areas of Thailand
Table 3
|
Pfdhfr haplotypesa
|
Amino acid position |
Prevalence (%) |
|
|
|
16 |
51 |
59 |
108 |
164 |
Total
n=170 |
Tak Province
n=81 |
Yala Province
n=89 |
|
Triple mutation |
A |
I |
R |
N |
I |
78 (45.9) |
6 (7.4) |
72 (80.9) |
|
|
Triple mutation |
A |
N |
R |
N |
L |
3 (1.8) |
3 (3.7) |
0 (0.0) |
|
|
Quadruple mutation |
A |
I |
R |
N |
L |
89 (52.4) |
72 (88.9) |
17 (19.1) |
|
|
Pfdhps haplotypesa
|
Amino acid position |
Prevalence (%) |
|
|
|
436 |
437 |
540 |
581 |
613 |
Total
n=170 |
Tak Province
n=81 |
Yala Province
n=89 |
|
|
Double mutation |
S |
G |
E |
A |
A |
2 (1.2) |
2 (2.5) |
0 (0.0) |
|
|
Double mutation |
S |
G |
K |
G |
A |
85 (50.0) |
11 (13.6) |
74 (83.1) |
|
|
Triple mutation |
A |
G |
E |
A |
A |
2 (1.2) |
2 (2.5) |
0 (0.0) |
|
|
Triple mutation |
A |
G |
K |
G |
A |
16 (9.4) |
1 (1.2) |
15 (16.9) |
|
|
Triple mutation |
S |
G |
E |
G |
A |
57 (33.5) |
57 (70.4) |
0 (0.0) |
|
|
Quadruple mutation |
A |
G |
E |
G |
A |
8 (4.7) |
8 (9.9) |
0 (0.0) |
Table 4Allele combinations of Plasmodium falciparum dihydrofolate reductase (Pfdhfr) and dihydropteroate synthase (Pfdhps) in 170 P. falciparum isolates from 2 endemic areas of Thailand
Table 4
|
Pfdhfr-Pfdhps allele combinations |
Prevalence (%) |
Total
n=170 |
Tak Province
n=81 |
Yala Province
n=89 |
|
AIRNI-AGEGA |
1 (0.6) |
1 (1.2) |
0 (0.0) |
|
AIRNI-AGKGA |
13 (7.6) |
0 (0.0) |
13 (14.6) |
|
AIRNI-SGEGA |
4 (2.4) |
4 (4.9) |
0 (0.0) |
|
AIRNI-SGKGA |
60 (35.3) |
1 (1.2) |
59 (66.3) |
|
AIRNL-AGEGA |
7 (4.1) |
7 (8.6) |
0 (0.0) |
|
AIRNL-AGKGA |
3 (1.8) |
1 (1.2) |
2 (2.2) |
|
AIRNL-SGEAA |
2 (1.2) |
2 (2.5) |
0 (0.0) |
|
AIRNL-SGEGA |
52 (30.6) |
52 (64.2) |
0 (0.0) |
|
AIRNL-SGKGA |
25 (14.7) |
10 (12.3) |
15 (16.9) |
|
ANRNL-AGEAA |
2 (1.2) |
2 (2.5) |
0 (0.0) |
|
ANRNL-SGEGA |
1 (0.6) |
1 (1.2) |
0 (0.0) |
References
- 1. Na-Bangchang K, Congpuong K. Current malaria status and distribution of drug resistance in East and Southeast Asia with special focus to Thailand. Tohoku J Exp Med 2007;211:99-113. https://doi.org/10.1620/tjem.211.99
- 2. Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR. Epidemiology of drug-resistant malaria. Lancet Infect Dis 2002;2:209-218. https://doi.org/10.1016/s1473-3099(02)00239-6
- 3. Noedl H, Socheat D, Satimai W. Artemisinin-resistant malaria in Asia. N Engl J Med 2009;361:540-541. https://doi.org/10.1056/NEJMc0900231
- 4. Harinasuta T, Suntharasamai P, Viravan C. Chloroquine resistant falciparum malaria in Thailand. Lancet 1965;286:657-660. https://doi.org/10.1016/s0140-6736(65)90395-8
- 5. Young MD, Contacos PG, Stitcher JE, Millar JW. Drug resistance in Plasmodium falciparum from Thailand. Am J Trop Med Hyg 1963;12:305-314. https://doi.org/10.4269/ajtmh.1963.12.305
- 6. Chin W, Bear DM, Colwell EJ, Kosakal S. A comparative evaluation of sulfalene-trimethoprim and sulphormethoxine-pyrimethamine against falciparum malaria in Thailand. Am J Trop Med Hyg 1973;22:308-312. https://doi.org/10.4269/ajtmh.1973.22.308
- 7. Johnson DE, Roendej P, Williams RG. Falciparum malaria acquired in the area of the Thai-Khmer border resistant to treatment with Fansidar. Am J Trop Med Hyg 1982;31:907-912. https://doi.org/10.4269/ajtmh.1982.31.907
- 8. Wongsrichanalai C, Sirichaisinthop J, Karwacki JJ, Congpuong K, Miller RS, Pang L, Thimasarn K. Drug resistant malaria on the Thai-Myanmar and Thai-Cambodian borders. Southeast Asian J Trop Med Public Health 2001;32:41-49.
- 9. WHO. Mekong Malaria Programme Malaria in the Greater Mekong Subregion: Regional and Country Profiles; World Health Organization; Geneva, Switzerland: 2008. http://www.whothailand.org/EN/Section3/Section113.htm
- 10. Reeder JC, Rieckmann KH, Genton B, Lorry K, Wines B, Cowman AF. Point mutations in the dihydrofolate reductase and dihydropteroate synthetase genes and in vitro susceptibility to pyrimethamine and cycloguanil of Plasmodium falciparum isolates from Papua New Guinea. Am J Trop Med Hyg 1996;55:209-213. https://doi.org/10.4269/ajtmh.1996.55.209
- 11. Basco LK, Eldin de Pecoulas P, Wilson CM, Le Bras J, Mazabraud A. Point mutations in the dihydrofolate reductase-thymidylate synthase gene and pyrimethamine and cycloguanil resistance in Plasmodium falciparum. MolBiochem Parasitol 1995;69:135-138. https://doi.org/10.1016/0166-6851(94)00207-4
- 12. Plowe CV, Cortese JF, Djimde A, Nwanyanwu OC, Watkins WM, Winstanley PA, Estrada-Franco JG, Mollinedo RE, Avila JC, Cespedes JL, Carter D, Doumbo OK. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine–sulfadoxine use and resistance. J Infect Dis 1997;176:1590-1596. https://doi.org/10.1086/514159
- 13. Babiker HA, Pringle SJ, Abdel-Muhsin A, Mackinnon M, Hunt P, Walliker D. High-level chloroquine resistance in Sudanese isolates of Plasmodium falciparum is associated with mutations in the chloroquine resistance transporter gene pfcrt and the multidrug resistance gene pfmdr1. J Infect Dis 2001;183:1535-1538. https://doi.org/10.1086/320195
- 14. Kublin JG, Cortese JF, Njunju EM, Mukadam RA, Wirima JJ, Kazembe PN, Djimdé AA, Kouriba B, Taylor TE, Plowe CV. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis 2003;187:1870-1875. https://doi.org/10.1086/375419
- 15. Mohammed A, Ndaro A, Kalinga A, Manjurano A, Mosha JF, Mosha DF, van Zwetselaar M, Koenderink JB, Mosha FW, Alifrangis M, Reyburn H, Roper C, Kavishe RA. Trends in chloroquine resistance marker, Pfcrt-K76T mutation ten years after chloroquine withdrawal in Tanzania. Malar J 2013;12:415. https://doi.org/10.1186/1475-2875-12-415
- 16. Mwai L, Ochong E, Abdirahman A, Kiara SM, Ward S, Kokwaro G, Sasi P, Marsh K, Borrmann S, Mackinnon M, Nzila A. Chloroquine resistance before and after its withdrawal in Kenya. Malaria J 2009;8:106. https://doi.org/10.1186/1475-2875-8-106
- 17. Wang X, Mu J, Li G, Chen P, Guo X, Fu L, Chen L, Su X, Wellems TE. Decreased prevalence of the Plasmodium falciparum chloroquine resistance transporter 76T marker associated with cessation of chloroquine use against P. falciparum malaria in Hainan, People’s Republic of China. Am J Trop Med Hyg 2005;72:410-414.
- 18. McCollum AM, Mueller K, Villegas L, Udhayakumar V, Escalante AA. Common origin and fixation of Plasmodium falciparum dhfr and dhps mutations associated with sulfadoxine-pyrimethamine resistance in a low-transmission area in South America. Antimicrob Agents Chemother 2007;51:2085-2091. https://doi.org/10.1128/AAC.01228-06
- 19. Hailemeskel E, Kassa M, Taddesse G, Mohammed H, Woyessa A, Tasew G, Sleshi M, Kebede A, Petros B. Prevalence of sulfadoxine-pyrimethamine resistance-associated mutations in dhfr and dhps genes of Plasmodium falciparum three years after SP withdrawal in Bahir Dar, Northwest Ethiopia. Acta Trop 2013;128:636-641. https://doi.org/10.1016/j.actatropica.2013.09.010
- 20. Pearce RJ, Ord R, Kaur H, Lupala C, Schellenberg J, Shirima K, Manzi F, Alonso P, Tanner M, Mshinda H, Roper C, Schellenberg D. A community-randomized evaluation of the effect of intermittent preventive treatment in infants on antimalarial drug resistance in southern Tanzania. J Infect Dis 2013;207:848-859. https://doi.org/10.1093/infdis/jis742
- 21. Raman J, Sharp B, Kleinschmidt I, Roper C, Streat E, Kelly V, Barnes KI. Differential effect of regional drug pressure on dihydrofolate reductase and dihydropteroate synthetase mutations in southern Mozambique. Am J Trop Med Hyg 2008;78:256-261. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3748784
- 22. Khim N, Bouchier C, Ekala MT, Incardona S, Lim P, Legrand E, Jambou R, Doung S, Puijalon OM, Fandeur T. Countrywide survey shows very high prevalence of Plasmodium falciparum multilocus resistance genotypes in Cambodia. Antimicrob Agents Chemother 2005;49:3147-3152. https://doi.org/10.1128/AAC.49.8.3147-3152.2005
- 23. McCollum AM, Mueller K, Villegas L, Udhayakumar V, Escalante AA. Common origin and fixation of Plasmodium falciparum dhfr and dhps mutations associated with sulfadoxine-pyrimethamine resistance in a low-transmission area in South America. Antimicrob Agents Chemother 2007;51:2085-2091. https://doi.org/10.1128/AAC.01228-06
- 24. Duraisingh MT, Curtis J, Warhurst DC. Plasmodium falciparum: detection of polymorphisms in the dihydrofolate reductase and dihydropteroate synthetase genes by PCR and restriction digestion. Exp Parasitol 1998;89:1-8. https://doi.org/10.1006/expr.1998.4274
- 25. Djimdé A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourté Y, Coulibaly D, Dicko A, Su XZ, Nomura T, Fidock DA, Wellems TE, Plowe CV. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med 2001;344:257-263. https://doi.org/10.1056/NEJM200101253440403
- 26. Maguire JD, AIKrisin Susanti, Sismadi P, Fryauff DJ, Baird JK. The T76 mutation in the pfcrt gene of Plasmodium falciparum and clinical chloroquine resistance phenotypes in Papua, Indonesia. Ann Trop Med Parasitol 2001;95:559-572. https://doi.org/10.1080/00034980120092516
- 27. Golassa L, Enweji N, Erko B, Aseffa A, Swedberg G. Detection of a substantial number of submicroscopic Plasmodium falciparum infections by polymerase chain reaction: a potential threat to malaria control and diagnosis in Ethiopia. Malar J 2013;12:352. https://doi.org/10.1186/1475-2875-12-352
- 28. Jiang T, Chen J, Fu H, Wu K, Yao Y, Eyi JUM, Matesa RA, Obono MMO, Du W, Tan H, Lin M, Li J. High prevalence of Pfdhfr–Pfdhps quadruple mutations associated with sulfadoxine–pyrimethamine resistance in Plasmodium falciparum isolates from Bioko Island, Equatorial Guinea. Malar J 2019;18:101. https://doi.org/10.1186/s12936-019-2734-x
- 29. Basuki S, Fitriah , Riyanto S, Budiono , Dachlan YP, Uemura H. Two novel mutations of pfdhps K540T and I588F, affecting sulphadoxine-pyrimethamine-resistant response in uncomplicated falciparum malaria at Banjar district, South Kalimantan Province, Indonesia. Malar J 2014;13:135. https://doi.org/10.1186/1475-2875-13-135
- 30. Lau TY, Sylvi M, William T. Mutational analysis of Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase genes in the interior division of Sabah, Malaysia. Malar J 2013;12:445. https://doi.org/10.1186/1475-2875-12-445
- 31. Zhao Y, Liu Z, Soe MT, Wang L, Soe TN, Wei H, Than A, Aung PL, Li Y, Zhang X, Hu Y, Wei H, Zhang Y, Burgess J, Siddiqui FA, Menezes L, Wang Q, Kyaw MP, Cao Y, Cui L. Genetic variations associated with drug resistance markers in asymptomatic Plasmodium falciparum infections in Myanmar. Genes 2019;10:692. https://doi.org/10.3390/genes10090692
- 32. Alam MT, Vinayak S, Congpuong K, Wongsrichanalai C, Satimai W, Slutsker L, Escalante AA, Barnwell JW, Udhayakumar V. Tracking origins and spread of sulfadoxine-resistant Plasmodium falciparum dhps alleles in Thailand. Antimicrob Agents Chemother 2011;155-164. https://doi.org/10.1128/AAC.00691-10
- 33. Sugaram R, Suwannasin K, Kunasol C, Mathema VB, Day NPJ, Sudathip P, Prempree P, Dondorp AM, Imwong M. Molecular characterization of Plasmodium falciparum antifolate resistance markers in Thailand between 2008 and 2016. Malar J 2020;19:107. https://doi.org/10.1186/s12936-020-03176-x
- 34. Imwong M, Jindakhad T, Kunasol C, Sutawong K, Vejakama P, Dondorp AM. An outbreak of artemisinin resistant falciparum malaria in Eastern Thailand. Sci Rep 2015;5:17412. https://doi.org/10.1038/srep17412
- 35. Krudsood S, Imwong M, Wilairatana P, Pukrittayakamee S, Nonprasert A, Snounou G, White NJ, Looareesuwan S. Artesunate–dapsone–proguanil treatment of falciparum malaria: genotypic determinants of therapeutic response. Trans R Soc Trop Med Hyg 2005;99:142-149. https://doi.org/10.1016/j.trstmh.2004.07.001
- 36. Setthaudom C, Tan-ariya P, Sitthichot N, Khositnithikul R, Suwandittakul N, Leelayoova S, Mungthin M. Role of Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes on in vitro chloroquine resistance in isolates of Plasmodium falciparum from Thailand. Am J Trop Med Hyg 2011;85:606-611. https://doi.org/10.4269/ajtmh.2011.11-0108
- 37. Holmgren G, Gil JP, Ferreira PM, Veiga MI, Obonyo CO, Björkman A. Amodiaquine resistant Plasmodium falciparum malaria in vivo is associated with selection of pfcrt 76T and pfmdr1 86Y. Infect Genet Evol 2006;6:309-314. https://doi.org/10.1016/j.meegid.2005.09.001
- 38. Chen N, Kyle DE, Pasay C, Fowler EV, Baker J, Peters JM, Cheng Q. pfcrt allelic types with two novel amino acid mutations in chloroquine-resistant Plasmodium falciparum isolates from the Philippines. Antimicrob Agents Chemother 2003;47:3500-3505. https://doi.org/10.1128/AAC.47.11.3500-3505.2003
- 39. Lopes D, Rungsihirunrat K, Nogueira F, Seugorn A, Gil JP, do Rosário VE, Cravo P. Molecular characterisation of drug-resistant Plasmodium falciparum from Thailand. Malaria J 2002;1:12. https://doi.org/10.1186/1475-2875-1-12
- 40. Congpuong K, Na Bangchang K, Mungthin M, Bualombai P, Wernsdorfer WH. Molecular epidemiology of drug resistance markers of Plasmodium falciparum malaria in Thailand. Trop Med Int Health 2005;8:717-722. https://doi.org/10.1111/j.1365-3156.2005.01450.x
- 41. Rungsihirunrat K, Chaijareonkul W, Seugorn A, Na-Bangchang K, Thaithong S. Association between chloroquine resistance phenotypes and point mutations in pfcrt and pfmdr1 in Plasmodium falciparum isolates from Thailand. Acta Trop 2009;109:37-40. https://doi.org/10.1016/j.actatropica.2008.09.011