Abstract
Bovine borreliosis, caused by Borrelia theileri which is transmitted via hard tick bites, is associated with mild clinical symptoms, such as fever, lethargy, hemoglobinuria, anorexia, and anemia. Borrelia theileri infects various animals, such as cattle, deer, horses, goats, sheep, and wild ruminants, in Africa, Australia, and South America. Notably, no case of B. theileri infection has been reported in Korean cattle to date. In this study, 101 blood samples were collected from a Korean indigenous cattle breed, among which 1.98% tested positive for B. theileri via nested PCR. The obtained sequences exhibited high homology with B. theileri strains identified in other regions. Phylogenetic analysis of 16S rRNA confirmed the B. theileri group affiliation; however, flagellin B sequences exhibited divergence, potentially due to regional evolutionary differences. This study provides the first molecular confirmation of B. theileri infection in Korean livestock. Further isolation and nucleotide sequence analyses are necessary to better understand the presence of B. theileri strains in cows in Korea.
-
Key words: Borrelia theileri, cattle, Korea, tick-borne pathogen
Borrelia species, which are mainly transmitted by tick bites, are divided into 2 groups; one group causing Lyme disease and the other invoking relapsing fever [
1]. Spirochetes in the Lyme disease group are associated with
B. afzelii,
B. garinii, and
B. burgdorferi, which are primarily transmitted by hard ticks. Recently, the relapsing fever group has been further subdivided into soft-tick-borne (associated with
B. coriaceae,
B. duttonii,
B. hermsii,
B. microtii, and
B. parkeri) and hard-tick-borne (associated with
B. lonstari,
B. miyamotoi, and
B. theileri) subgroups based on their vectors [
2].
B. theileri infects various animals, such as cattle, deer, horses, goats, sheep, and wild ruminants, in Africa, Australia, and South America [
2–
8]. Bovine borreliosis is caused by
B. theileri that is transmitted via the bite of
Rhipicephalus species and is associated with mild clinical symptoms, such as fever, lethargy, hemoglobinuria, anorexia, and anemia [
2,
4,
9].
B. theileri DNA has been reported in raccoon dogs in southern part of Korean peninsula, while
B. theileri-like
Borrelia species have been reported in ticks obtained from goats in northern part of Korean peninsula [
10,
11]. However, no case of
B. theileri infection has been reported in Korean cattle. This study aimed to investigate the presence of
B. theileri in cows in Korea using nested PCR.
Blood samples were collected from the jugular veins of cattle in Korea into Vacutainer sodium heparin tubes (BD Biosciences, Franklin Lakes, NJ, USA) from October to December 2022. DNA extraction from the blood samples was performed using the FavorPrep Blood/Cultured Cell Genomic DNA Extraction Mini Kit (Favorgen Biotech, Ping Tung, Taiwan), following the manufacturer’s protocol. Conventional and nested PCRs were conducted using primers specific for 16S rRNA, outer surface protein A (
ospA), outer surface protein C (
ospC), and flagellin B (
flaB) genes of
Borrelia spp., as previously described [
12–
15]. The PCR reaction mixture consisted of 10–100 ng of genomic DNA for the first PCR, 1 μl of the first PCR product for the nested PCR, 10 pmoles of primers, 10×PCR buffer (TransGen Biotech, Beijing, China), 2.5 mm deoxyribonucleotide triphosphates (TransGen Biotech), and 1U of Taq polymerase (TransGen Biotech). Both the first and nested PCRs were performed in a total volume of 25 μl using a MiniAmp Thermal Cycler (Thermo Fisher Scientific, Waltham, MA, USA). The resulting PCR products were separated on a 1.5% agarose gel and were post-stained with ethidium bromide (Biosesang, Seongnam, Korea). Unfortunately, PCR products of
ospA and
ospC were not amplified, whereas only those of 16S rRNA and
flaB were successfully amplified. The amplified PCR products were purified using the FavorPrep GEL/PCR Purification Kit (Favorgen Biotech) and cloned using the pLUG-Prime TA-cloning Vector Kit II (iNtRON Biotechnology, Seongnam, Korea). Ten colonies per sample were selected, and plasmid DNAs for sequencing were purified using the FavorPrep Plasmid DNA Extraction Mini Kit (Favorgen Biotech), according to the manufacturer’s instructions. Purified recombinant plasmid DNAs were sequenced using an automatic sequencer (3730xl Capillary DNA Analyzer; Applied Biosystems, Foster City, CA, USA). The obtained sequences were analyzed using Chromas software (Ver 2.66). To compare the sequences generated from this study with those in the GenBank database, the sequences were aligned using Clustal X (Ver 2.1) and analyzed using MEGA 7. Phylogenetic trees were constructed using the maximum likelihood method based on the Tamura-Nei model, and the dataset was resampled 1,000 times to generate bootstrap values.
In total, 101 blood samples were collected from grazing Hanwoo cattle, which is a breed of cattle indigenous to Korea, from 2 provinces (Chungnam,
n=67; Gyeongnam,
n=34). Two samples tested positive for
B. theileri (1.98%).
B. theileri 16S rRNA and
flaB were detected using nested PCR. The amplicon sizes obtained were 507 and 323 bp, respectively. The obtained
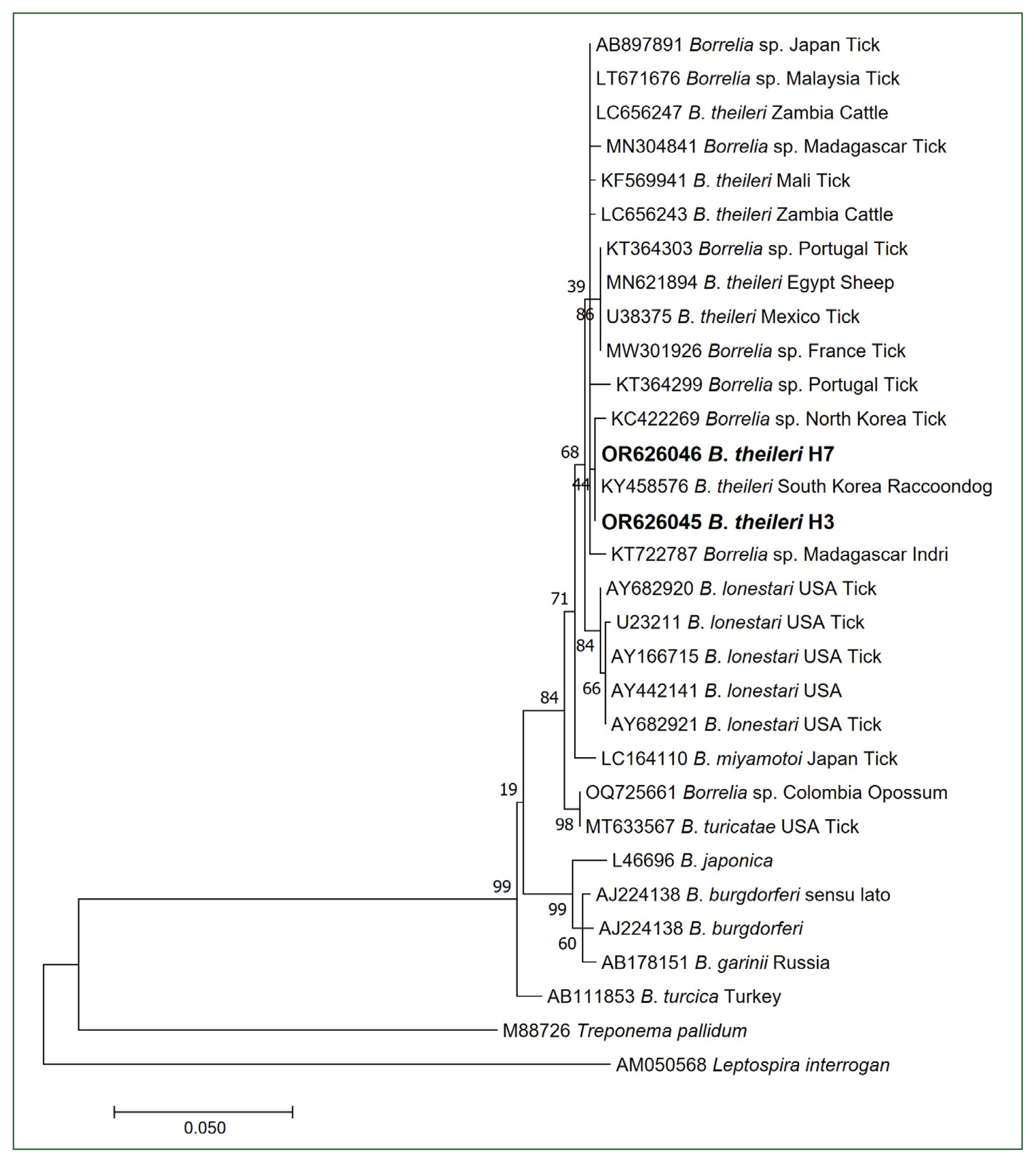
B. theileri 16S rRNA sequences, H3 (OR626045) and H7 (OR626046), were identical to each other and showed 100% homology with a
B. theileri sequence (KY458576) obtained from a raccoon dog in Korea. Additionally, these
B. theileri sequences exhibited 99.86% homology with a
Borrelia sp. sequence (KC422269) isolated from ticks in North Korea, 99.58% homology with a
B. theileri sequence (MN621894) isolated from sheep in Egypt, and 99.58% homology with a
B. theileri sequence (U38375) isolated from ticks in Mexico. Based on phylogenetic analysis, the 16S rRNA sequences (H3 and H7) were unequivocally identified as belonging to the
B. theileri group (
Fig. 1). As the 16S rRNA gene is highly conserved, we conducted additional PCRs using primer sets that target the
Borrelia flagellin gene. The obtained
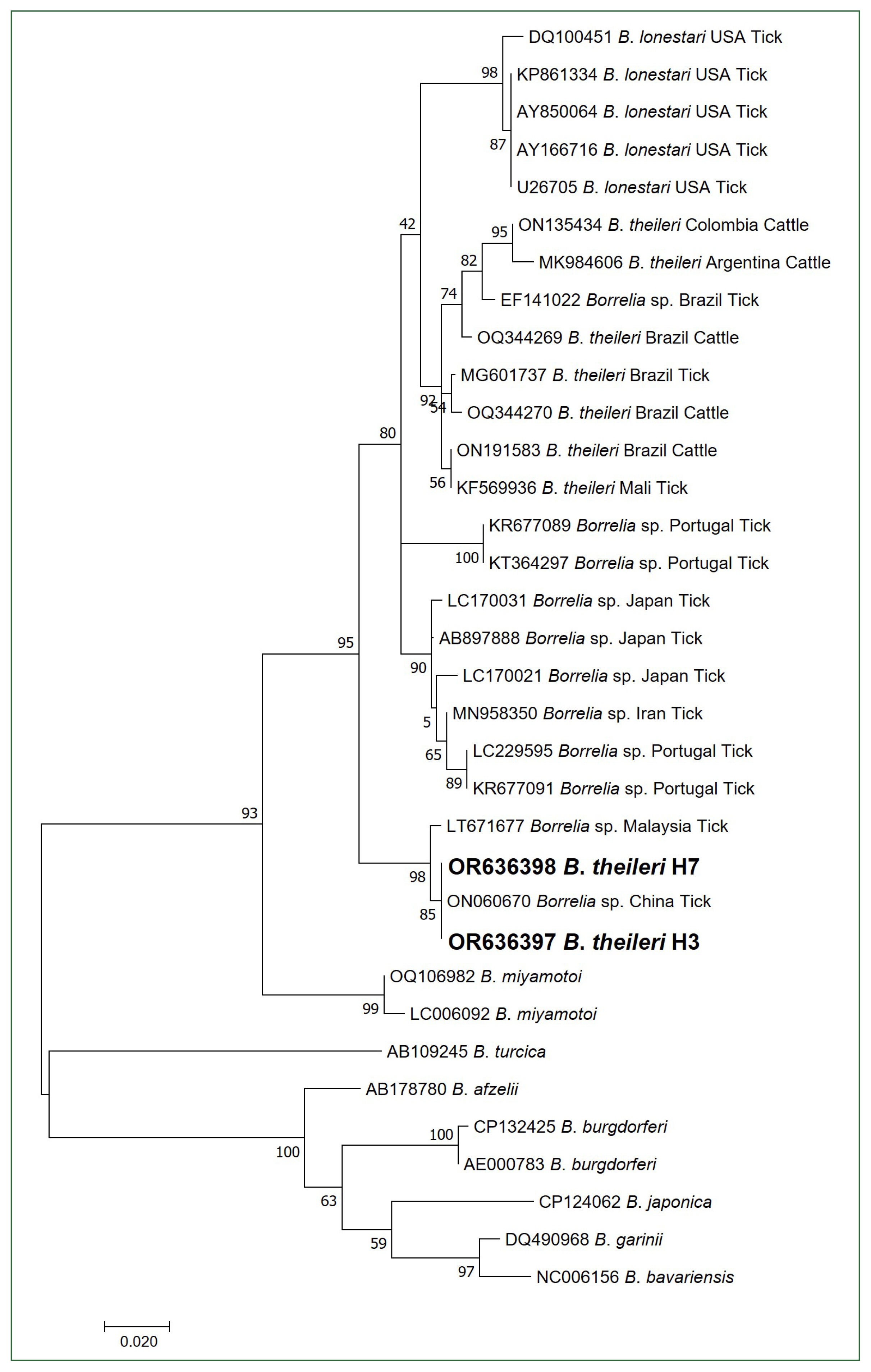
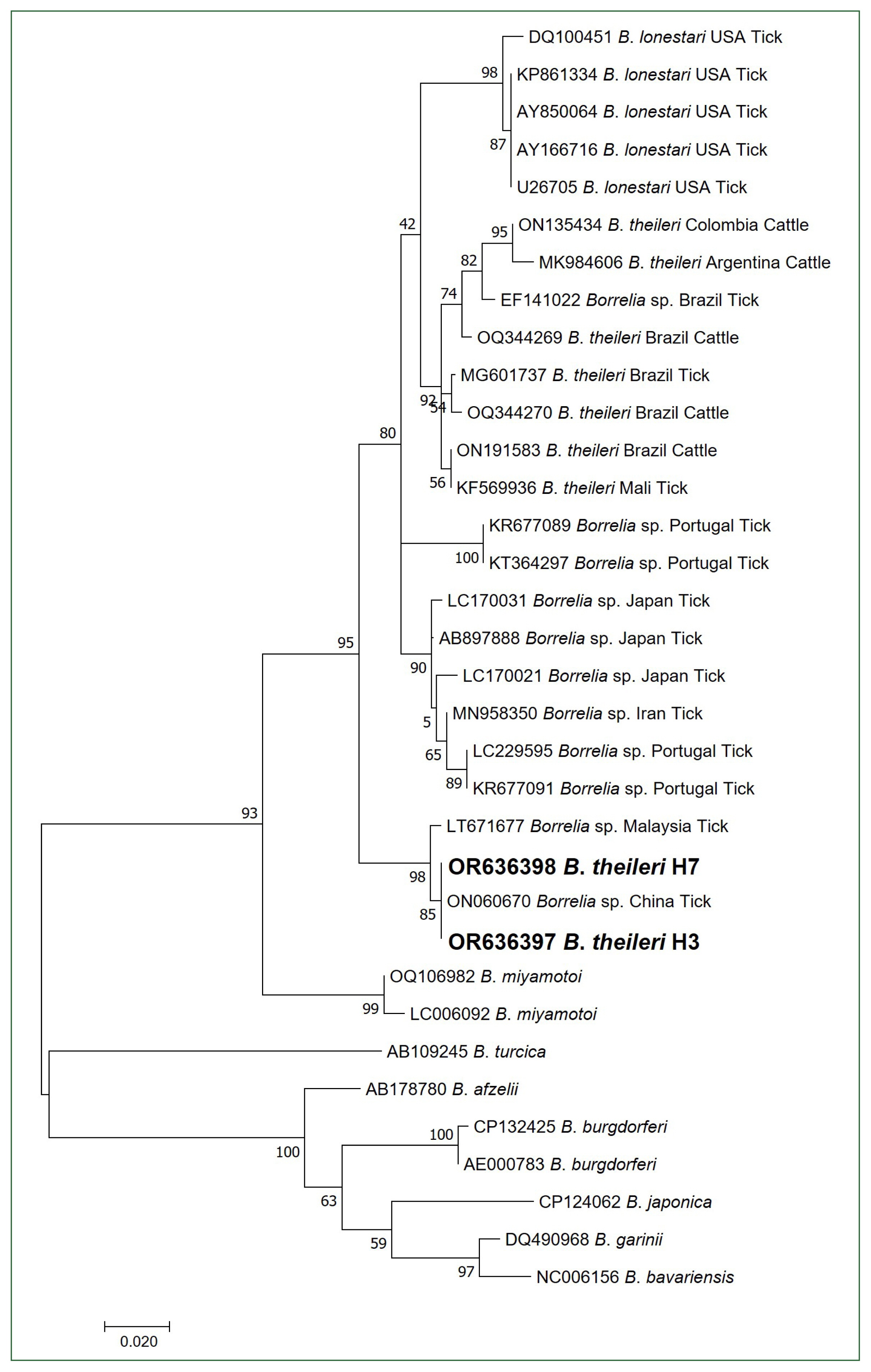
B. theileri flaB sequences, H3 (OR636397) and H7 (OR636398), were identical to each other and showed 100% homology with a
Borrelia sp. sequence (ON060670) isolated from ticks in China, 99.38% homology with a
Borrelia sp. sequence (LT671677) isolated from ticks in Malaysia, 95.67% homology with a
B. theileri sequence (ON191583) isolated from cattle in Brazil, 95.36% homology with a
Borrelia sp. sequence (AB897888) isolated from ticks in Japan, 95.36% homology with a
B. theileri sequence (MG601737) isolated from ticks in Brazil, 95.05% homology with a
B. lonstari sequence (AY850064) isolated from ticks in the USA, and 94.74% homology with a
B. theileri sequence (ON135434) isolated from cattle in Colombia.
Unlike the 16S rRNA phylogenetic analysis, the obtained
flaB sequences were not included in the
B. theileri group and clustered with an unidentified
Borrelia species group (
Fig. 2). Several reasons could explain these observations. First, the number of registered
flaB sequences for
B. theileri in GenBank was notably low, suggesting the inadequate analysis of nucleotide variations among
Borrelia species. Second, although speculating solely based on the current results is not completely accurate, potential evolutionary differences in flagellin genes may be observed in
Borrelia species found in Asian regions compared to the known
B. theileri species found in other regions. The primary vectors of
B. theileri are
Rhipicephalus species, which inhabit Africa, Australia, South America, and Southern Asia [
2]. Moreover,
Rhipicephalus sanguineus has been identified; however, it is a rare tick species in Korea [
16]. In contrast,
Haemaphysalis species are the predominant hard tick species in Korea [
10,
17]. Moreover, Hanwoo is a breed of cattle unique to Korea. These environmental differences between ticks and their hosts may affect the presence of
B. theileri.
Currently, only 4 B. theileri sequences from Korea, including those presented in this study, are available in the GenBank database for phylogenetic analyses. Although we attempted to isolate this bacterium for precise species analysis, our efforts were unsuccessful. Nevertheless, this study is the first to use molecular techniques to confirm the presence of B. theileri in domestic livestock of Korea. However, further research is necessary to confirm the characteristics of B. theileri strains in Korea via B. theileri isolation and nucleotide sequence analyses. Additionally, future studies should investigate the presence of B. theileri in the predominant hard tick species (Haemaphysalis species) of Korea.
Notes
-
The authors declare no conflicts of interest related to this study.
-
Conceptualization: Hyung HJ, Lee KJ, Kang JG
Data curation: Hyung HJ, Choi YS, Park J
Formal analysis: Hyung HJ, Choi YS
Funding acquisition: Kang JG
Investigation: Hyung HJ, Choi YS, Park J
Methodology: Hyung HJ, Choi YS
Project administration: Kang JG
Resources: Park J, Lee KJ
Software: Kang JG
Supervision: Lee KJ, Kang JG
Visualization: Hyung HJ, Kang JG
Writing – original draft: Hyung HJ, Kang JG
Writing – review & editing: Hyung HJ, Choi YS, Park J, Lee KJ, Kang JG
Acknowledgment
This paper was supported by research funds for newly appointed professors of Jeonbuk National University in 2020 and by the National Institutes of Health Research Project (Project No. 2022ER210801).
Fig. 1Phylogenetic tree of Borrelia species based on partial 16S rRNA sequences (507 bp). Bold letters indicate the Borrelia theileri sequences obtained from cattle in this study. The scale bar depicts the nucleotide substitution per position. Numbers at the nodes indicate the proportions of 1,000 bootstrap iterations that support the topology shown.
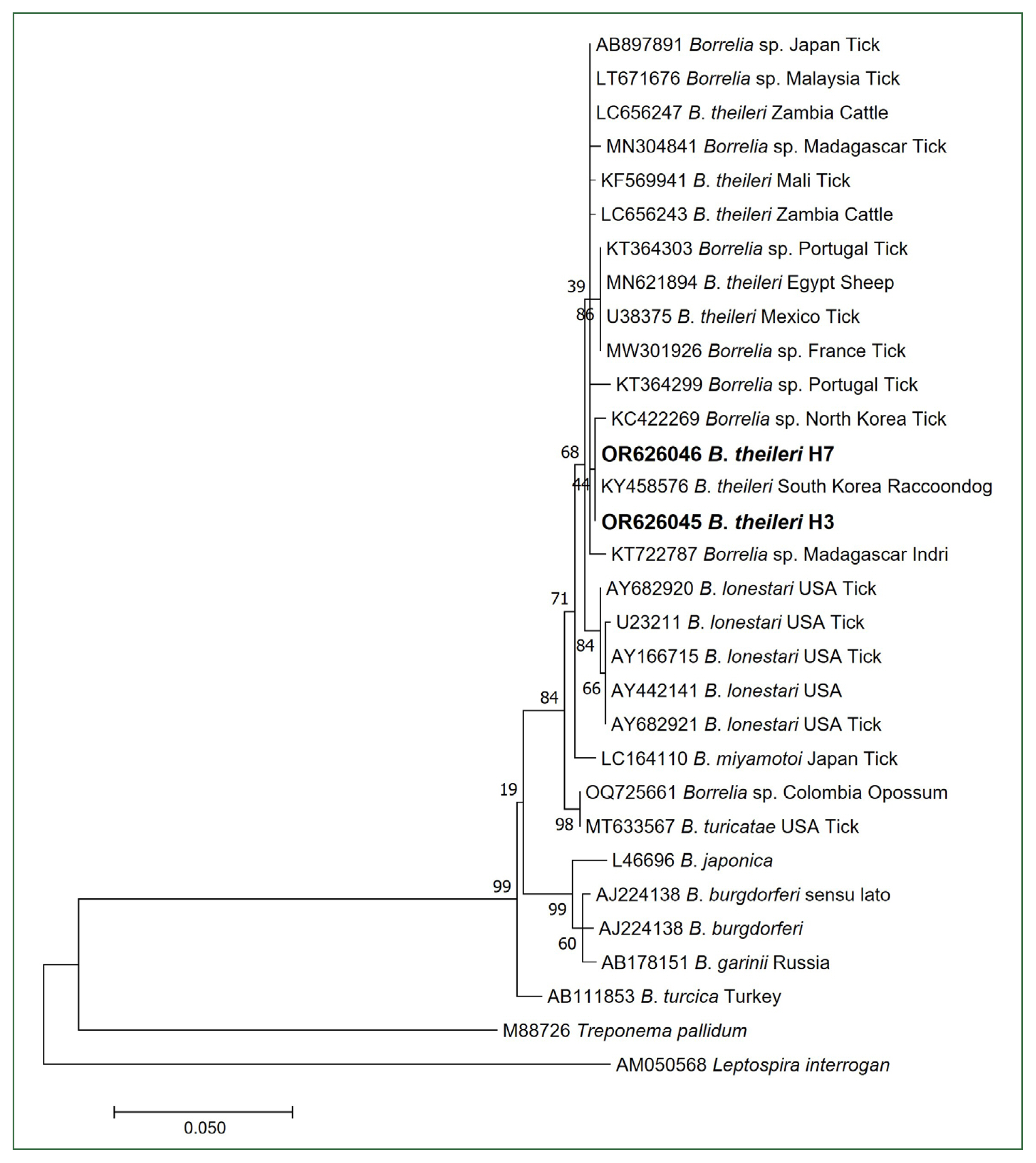
Fig. 2Phylogenetic tree of Borrelia species based on partial flagellin B gene sequences (323 bp). Bold letters indicate the B. theileri sequences obtained from cattle in this study. The scale bar depicts the nucleotide substitution per position. Numbers at nodes indicate the proportions of 1,000 bootstrap iterations that support the topology shown.
References
- 1. Margos G, Vollmer SA, Ogden NH, Fish D. Population genetics, taxonomy, phylogeny and evolution of Borrelia burgdorferi sensu lato. Infect Genet Evol 2011;11(7):1545-1563. https://doi.org/10.1016/j.meegid.2011.07.022
- 2. Trevisan G, Cinco M, Trevisini S, di Meo N, Ruscio M, et al. Borreliae Part 2: Borrelia Relapsing Fever Group and Unclassified Borrelia. Biology (Basel) 2021;10(11):1117. https://doi.org/10.3390/biology10111117
- 3. Theiler A, Bruce D. Transmission and inoculability of Spirillum theileri (Laveran). Proc R Soc Lond 1905;76(512):504-506. https://doi.org/10.1098/rspb.1905.0043
- 4. Callow LL. Observations on tick-transmitted spirochaetes of cattle in Australia and South Africa. Br Vet J 1967;123(11):492-497. https://doi.org/10.1016/S0007-1935(17)39704-X
- 5. Aouadi A, Leulmi H, Boucheikhchoukh M, Benakhla A, Raoult D, et al. Molecular evidence of tick-borne hemoprotozoan-parasites (Theileria ovis and Babesia ovis) and bacteria in ticks and blood from small ruminants in Northern Algeria. Comp Immunol Microbiol Infect Dis 2017;50:34-39. https://doi.org/10.1016/j.cimid.2016.11.008
- 6. Abdullah HHAM, Amanzougaghene N, Dahmana H, Louni M, Raoult D, et al. Multiple vector-borne pathogens of domestic animals in Egypt. PLoS Negl Trop Dis 2021;15(9):e0009767. https://doi.org/10.1371/journal.pntd.0009767
- 7. Paula WVF, Neves LC, de Paula LGF, Serpa MCA, de Oliveira FP, et al. First molecular detection of Borrelia theileri subclinical infection in a cow from Brazil. Vet Res Commun 2023;47(2):963-967. https://doi.org/10.1007/s11259-022-10020-x
- 8. Qiu Y, Squarre D, Nakamura Y, Lau ACC, Moonga LC, et al. Evidence of Borrelia theileri in wild and domestic animals in the Kafue ecosystem of Zambia. Microorganisms 2021;9(11):2405. https://doi.org/10.3390/microorganisms9112405
- 9. Trees AJ. The transmission of Borrelia theileri by Boophilus annulatus (Say, 1821). Trop Anim Health Prod 1978;10(2):93-94. https://doi.org/10.1007/BF02235315
- 10. Kang JG, Ko S, Smith WB, Kim HC, Lee IY, et al. Prevalence of Anaplasma, Bartonella and Borrelia species in Haemaphysalis longicornis collected from goats in North Korea. J Vet Sci 2016;17(2):207-216. https://doi.org/10.4142/jvs.2016.17.2.207
- 11. Kang JG, Chae JB, Cho YK, Jo YS, Shin NS, et al. Molecular detection of Anaplasma, Bartonella, and Borrelia theileri in raccoon dogs (Nyctereutes procyonoides) in Korea. Am J Trop Med Hyg 2018;98(4):1061-1068. https://doi.org/10.4269/ajtmh.17-0380
- 12. Takano A, Fujita H, Kadosaka T, Konnai S, Tajima T, et al. Characterization of reptile-associated Borrelia sp. in the vector tick, Amblyomma geoemydae, and its association with Lyme disease and relapsing fever Borrelia spp. Environ Microbiol Rep 2011;3(5):632-637. https://doi.org/10.1111/j.1758-2229.2011.00280.x
- 13. Kang JG, Kim HC, Choi CY, Nam HY, Chae HY, et al. Molecular detection of Anaplasma, Bartonella, and Borrelia species in ticks collected from migratory birds from Hong-do Island, Republic of Korea. Vector Borne Zoonotic Dis 2013;13(4):215-225. https://doi.org/10.1089/vbz.2012.1149
- 14. Priem S, Rittig MG, Kamradt T, Burmester GR, Krause A. An optimized PCR leads to rapid and highly sensitive detection of Borrelia burgdorferi in patients with lyme borreliosis. J Clin Microbiol 1997;35(3):685-690. https://doi.org/10.1128/jcm.35.3.685-690.1997
- 15. Marti Ras N, Postic D, Foretz M, Baranton G. Borrelia burgdorferi sensu stricto, a bacterial species “made in the U.S.A.”? Int J Syst Bacteriol 1997;47(4):1112-1117. https://doi.org/10.1099/00207713-47-4-1112
- 16. Kang YB. Rhipicephalus has been identified in Korea. Rhipicephalus sanguineus (Latreille 1806); a new record of male tick identified with scanning electron microscopy in Korea. Korean J Vet Res 1984;24(2):201-212.
- 17. Jo YS, Kang JG, Chae JB, Cho YK, Shin JH, et al. Prevalence of severe fever with thrombocytopenia syndrome virus in ticks collected from national parks in Korea. Vector Borne Zoonotic Dis 2019;19(4):284-289. https://doi.org/10.1089/vbz.2018.2338
Citations
Citations to this article as recorded by

- Nationwide Geographical and Temporal Distribution of Tick-Borne Diseases in Korean Water Deer (Hydropotes inermis argyropus)
Beoul Kim, Su-Jin Chae, You-Jeong Lee, Haksub Shin, Sunmin Kwak, Hyesung Jeong, Suwoong Lee, Dongmi Kwak, Min-Goo Seo
Animals.2025; 15(10): 1499. CrossRef - Circulation of tick-borne pathogens in wildlife of the Republic of Korea
Hye-ryung Byun, Seong-Ryeong Ji, Jun-Gu Kang, Chang-Yong Choi, Ki-Jeong Na, Jong-Taek Kim, Joon-Seok Chae
One Health.2024; 19: 100913. CrossRef