Abstract
Toxoplasma gondii primarily invades the central nervous system, causing latent infections. Cysts persist in the host for life and there is currently no effective treatment. T. gondii infects human hosts through contaminated meat, invading the intestinal tissue and leading to changes in the number and composition of the gut microbiota. Since probiotic ingestion modulates intestinal microbiota changes, we hypothesized that intestinal microbiota dysbiosis caused by T. gondii infection would be restored following probiotic supplementation. To this end, we orally infected C57BL/6 mice with 10 T. gondii cysts and administered supplemental probiotics daily. We analyzed the levels of T. gondii B1 gene DNA, indicative of T. gondii infection, in brain tissue. We investigated alterations in the gut microbiota composition and functional pathways between the probiotic and non-probiotic treatment groups via next-generation sequencing analysis of each fecal sample. The infection level in the probiotic-treated group was significantly reduced after 4 weeks (P<0.05). Probiotic supplementation notably changed the gut microbiota after 2 weeks of infection, increasing the relative abundance of Intestinimonas massiliensis and Lawsonibacter asaccharolyticus. Probiotic supplements appear to modulate the gut microbiota, activating functional pathways involved in intestinal short-chain fatty acid production and strengthening the intestinal barrier, thereby impeding T. gondii infection and subsequent proliferation. Our findings provide valuable insights into T. gondii infection control and future study directions.
-
Key words: Toxoplasma gondii, probiotics, microbiota, next-generation sequencing
Introduction
Toxoplasma gondii is a prominent foodborne pathogen, infecting approximately one-third of humans worldwide [
1]. It is commonly acquired by ingesting cysts present in undercooked meat or contaminated water [
2]. Upon host entry,
T. gondii undergoes 2 life cycle stages: A rapidly dividing tachyzoite stage at the beginning of the infection and a slow-growing, latent bradyzoite stage, with cyst formation in various host tissues. When the bradyzoites are released from the cysts, they develop into tachyzoites, infecting the surrounding tissues and spreading to other organs [
3]. After initially infecting intestinal epithelial cells,
T. gondii circulates in the host’s bloodstream and forms localized cysts [
4]. Typically,
T. gondii invades the central nervous system, causing lifelong latent infections that are incurable [
5]. If the immune system becomes compromised,
T. gondii reactivates from its latent state, leading to the development of toxoplasmic encephalitis [
5] or ocular disease [
6]. Furthermore,
T. gondii invasion of the brain potentially contributes to the development of various psychiatric disorders, including behavioral and personality changes [
7].
Novel treatment options are crucial because current treatments for active
T. gondii tachyzoites are ineffective, require long-term administration, and are associated with severe side effects [
8]. Furthermore, there is no effective agent against latent bradyzoites [
7]. Since no cure exists, inhibiting penetration and reducing the infection severity in the gut, which serves as the first line of defense, may be an effective management strategy. This study aimed to determine the potential role of probiotics in inhibiting
T. gondii infection. Ingested
T. gondii first invades the small intestine, where it penetrates the intestinal barrier, travels deeper, and spreads to distant organs via lymphatic and blood vessels [
5]. During the initial stage of
T. gondii infection in the intestinal tissues, the parasite continuously secretes various factors, resulting in complex interactions involving the gut microbiota,
T. gondii, and the mucosal immune system. This leads to rapid changes in the number and composition of the gut microbiota [
9,
10], influencing the onset and severity of various diseases and affecting the intestinal mucosa and immune function [
11]. Several studies have demonstrated that
T. gondii and other parasites, such as
Blastocystis spp. and
Cryptosporidium parvum, affect gut microbiota composition. Conversely, infection progression is regulated by gut microbiota composition [
10]. A recent study where
T. gondii-infected mice were fed koumiss (a fermented dairy product) induced gut microbiota changes and reduced the number of
T. gondii cysts in the brain at 42 days after infection [
12]. However, data on the role of gut microbiota composition in
T. gondii infection are lacking.
The health benefits of probiotics have been established, including gut microbiota and immune system modulation [
13]. Various studies have investigated treating infectious diseases by improving the gut microbiota using probiotics [
14]. From classical probiotics, such as
Lactobacillus and
Bifidobacterium, to newer and diverse probiotic strains, probiotics have been shown to alter the gut microbiota, thereby improving metabolism, the epithelial barrier, and immune response [
15].
It is expected that probiotics may reduce
T. gondii infection in mice by inducing favorable changes in the gut microbiota. This study analyzed changes in the gut microbiota of
T. gondii-infected mice that received supplemental probiotics in a controlled environment. We used a 21-strain probiotic primarily comprising lactic acid bacteria, including the most widely used probiotic strains [
16].
Materials and Methods
Ethics statement
All animal experiments were performed at the Korea Food Research Institute after protocol review and approval by the Institutional Animal Care and Use Committee (approval no: KFRI-M-23028).
Mouse maintenance and parasite infection
We purchased 6-week-old C57BL/6 male mice from Orient Bio (Gyeonggi-do, Korea). They were housed in the Experimental Animal Research Building of the Korea Food Research Institute and acclimated for 1 week. Groups of 3 or 2 mice were housed in individually ventilated cages (GM500; Tecniplast, Italy) with access to food and reverse osmosis water ad libitum. The quarantine and rearing rooms were maintained at 23±1°C, 55±5% humidity, with a 12/12 light/dark cycle, 300 lux illumination, and <60 dB noise based on laboratory animal husbandry standards. At the end of the acclimation period, the mice were divided into a control group (n=5 for each group after 0, 2, and 4 weeks of infection), an infected group (Tg; n=5 for each group after 0, 2, and 4 weeks of infection), and a probiotics group (Tg+PB; n=5 for each group after 0, 2, and 4 weeks of infection). First, the Tg and Tg+PB groups were orally infected with one dose of 100 μl sterile PBS containing 10 T. gondii ME49 cysts, and the control group received the same amount of sterile phosphate-buffered saline alone. Following infection, the Tg+PB group was orally administered 100 μl of saline solution containing dissolved probiotic powder (1×105 colony-forming units), and the other groups received the same amount of saline solution alone. The probiotics used in this study were commercially available and purchased from Huons Natural Co. (Jecheon, Korea). Before the experiment, the manufacturing process sheet, certificate of analysis, and manufacturing production report were thoroughly reviewed to ensure the integrity and quality of the ingredients (Lactiplantibacillus plantarum, Lacticaseibacillus rhamnosus, Lacticaseibacillus casei, Lacticaseibacillus paracasei, Lactobacillus acidophilus, Lactobacillus gasseri, Lactobacillus helveticus, Lactobacillus delbrueckii subsp. bulgaricus, Ligilactobacillus salivarius, Bifidobacterium breve, B. bifidum, B longum, B. animalis subsp. lactis, B. infantis, Streptococcus thermophilus, Lactococcus lactis subsp. lactis, Limosilactobacillus fermentum, Limosilactobacillus reuteri, Leuconostoc citreum, Leuconostoc kimchii, and Levilactobacillus brevis). The T. gondii strain ME49 cysts used to infect the mice in our experiment had been collected from the brain tissue of C57BL/6 mice after 6 weeks of infection.
Fecal and brain sample collection
After anesthetizing the mice with isoflurane, blood was collected by retro-orbital sampling using a capillary tube (Kimble Chase Life Science, Vineland, NJ, USA). The mice were sacrificed, after which the skull was opened. The entire brain was removed and homogenized. No perfusion was performed during brain harvesting. The fecal samples were collected from isolated intestine at the same time. Brain homogenate and fecal samples were stored at −80°C.
Quantitative PCR (qPCR) and statistical analysis of T. gondii-specific B1 gene
qPCR was performed using a CFX96 Real PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA) and TOPreal SYBR Green qPCR 2×PreMIX (Enzynomics, Daejeon, Korea). Briefly, total genomic DNA was isolated from the brain tissue homogenates using a DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) per the manufacturer’s protocol. The isolated genomic DNA underwent qPCR to amplify the T. gondii-specific B1 gene, and Gapdh was used as the reference gene. Statistical analyses were performed using GraphPad Prism 5 software (GraphPad Software, La Jolla, CA, USA). Data were expressed as the mean±SD. ANOVA was performed to evaluate differences between the experimental groups. P-values of <0.05 were considered statistically significant.
Fecal DNA extraction, library construction, and sequencing
DNA was extracted from fecal samples using a DNeasy PowerSoil Pro Kit (Qiagen) per the manufacturer’s protocol. Next, sequencing libraries were constructed according to 16S Metagenomic Sequencing Library protocols (Illumina, San Diego, CA, USA) to amplify the V3 and V4 regions of the gut microbiota. Each amplification reaction contained 5 ng of genomic DNA, 5×reaction buffer, 1 mM dNTP mix, 500 nM each of universal forward and reverse primers, and Herculase II fusion DNA polymerase (Agilent Technologies, Santa Clara, CA, USA). The primary PCR cycling conditions were 95°C for 3 min; 25 cycles of 95°C for 30 sec, 30°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec; and final extension at 72°C for 5 min. The primer sequences were as follows: 5′-tcgtcggcagcgtcagatggtgtaagagacagcctacggnggcwgcag-3′ (forward) and 5′-gtctcggtgtgctcggagatggtgtaagagacaggactachvggggtatctaatcc-3′ (reverse). The primary PCR product was purified with AMPure beads (Agencourt Bioscience, Beverly, MA, USA), and 10 μl of the purified product underwent a second round of PCR amplification using Nextera XT Indexed Primers (Illumina) to construct the final library. The PCR conditions were identical to those of the primary PCR except that 10 instead of 25 amplification cycles were performed. The PCR products were purified using AMPure beads, quantified using KAPA Library Quantification Kits for Illumina platforms according to the qPCR Quantification Protocol Guide (KAPA BioSystems, Wilmington, MA, USA), and validated using TapeStation D1000 ScreenTape (Agilent Technologies, Waldbronn, Germany). Finally, the samples were sequenced on a MiSeq platform (Illumina).
Preparation of amplicon sequence variants (ASVs) and comparative analysis of microbiota
After the DNA sequences were read using an Illumina MiSeq platform, each sample was separated by index sequence, and paired-end FASTQ files were generated. Adapters and primer sequences were removed using Cutadapt v3.2, and possible errors in sequence reading were corrected and excluded using the DADA2 package v1.18.0 in R v4.0.3 (R Core Team (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria) (excluding sequences with expected errors >2 for paired-end reads). After removing noise from the processed data on a sample-by-sample basis, the corrected sequences were merged to restore accurate DNA sequence information. Abnormal sequences (chimeras) were removed, and each unique sequence (ASV) was identified, with sequences <350 bp excluded from the analysis. QIIME v1.9 was used to normalize all samples to the same reference (read count of the sample with the lowest read count).
Analysis and statistical analysis of biological information
We searched the NCBI 16S Microbial Database for each ASV sequence using BLAST+ v2.9.0 and assigned taxonomic information according to the organism with the highest sequence similarity. Taxonomic information was not assigned if the query coverage of the best hit was <85% or if the sequence identity was <85%. QIIME v1.9 was used to perform comparative analyses using ASV abundance and taxonomy information. To enable detailed comparison of microbial communities between groups, taxonomy bar plots of mean relative abundance (%) were generated per group. The overall sample mean relative abundance value was used as a sorting reference. Taxa that were not in the top 15 results or those with a total sample mean abundance value <1% were lumped into the “other” taxon, placed at the top of the bar plot. Analysis of similarities (ANOSIM) was performed to assess the significance of differences between groups. In this analysis, the
R-value ranges from −1 to 1, with 1 indicating less similarity between groups, 0 indicating equal between- and within-group similarities, and −1 indicating within-group similarity lower than between-group similarity. Principal coordinates analysis (PCoA) was performed to investigate between-group differences based on weighted and unweighted UniFrac distances, and significant differences were checked using ANOSIM. For linear discriminant analysis effect size (LEfSe) analysis, data (taxon) with significant differences between the comparison groups were selected using the Kruskal–Wallis method based on the taxonomic assignment results. Then, the linear discriminant analysis score was calculated for the selected data. Only data with significant differences between groups were retained, from which only scores ≥2.0 were selected [
17]. MetaCyc metabolic pathways were also predicted per group to analyze the altered gut environment using PICRUSt2. After examining the microbial community, the functional abundance, with standardized ASV abundance after correction, was estimated using the 16S rRNA gene copy number database function in PICRUSt and expressed as pathway classification information. To increase the reliability of functional predictions, ASVs with nearest sequenced taxon index values >2 were excluded from the PICRUSt2 analysis [
18]. The functions were visualized by group using ggplot v3.3.2. The significance of the predicted increase in pathways involved in short-chain fatty acid (SCFA) production was assessed using
t-tests.
Results
Quantitative alterations in B1 gene DNA in the brain tissue of T. gondii-infected mice receiving supplemental probiotics
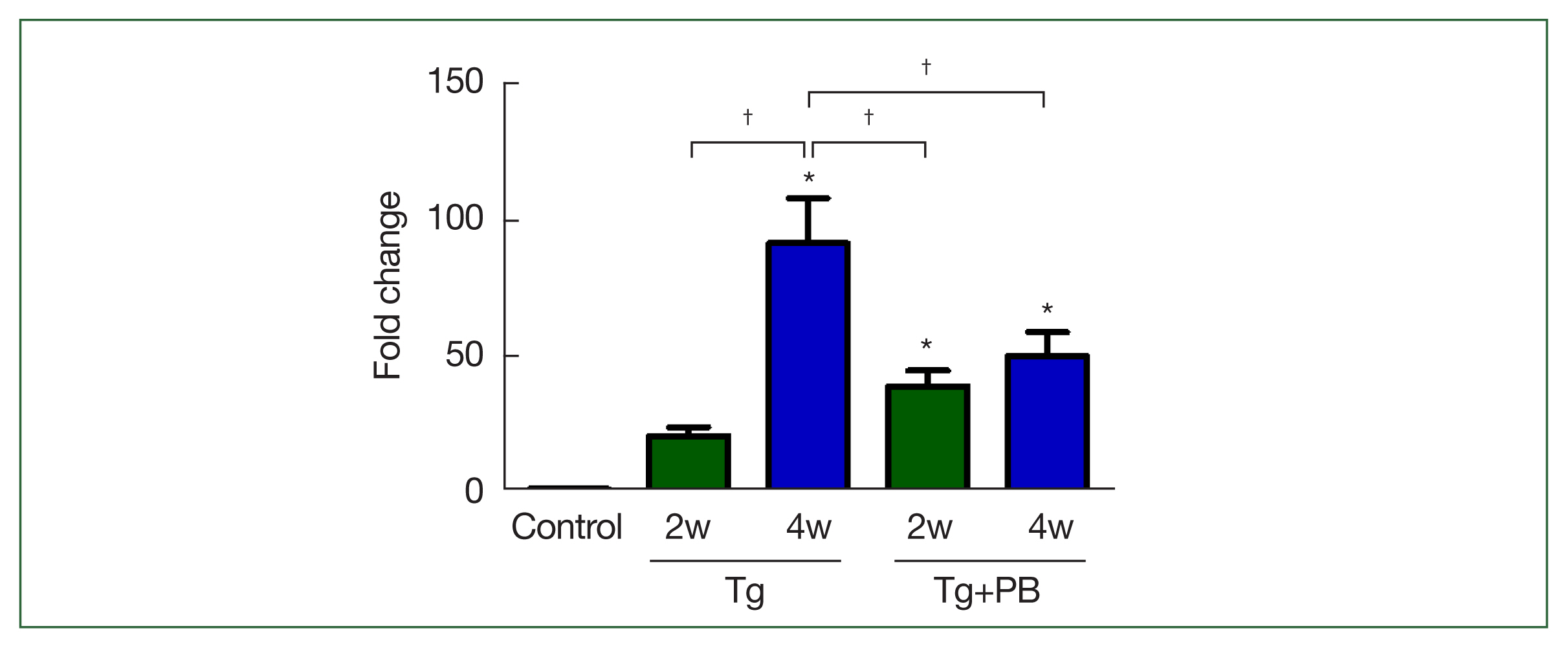
The inhibitory effect of probiotics on
T. gondii infection in mice was assessed after 2 and 4 weeks of infection. Unexpectedly, the B1 gene level in the Tg+PB group was significantly higher compared with the Tg group after 2 weeks of infection (
P<0.01). However, after 4 weeks of infection, the B1 gene level in the Tg+PB group was significantly reduced compared with the Tg group (
P<0.05) (
Fig. 1). Conversely, the B1 gene level in the Tg+PB group did not significantly increase between 2 and 4 weeks after infection, confirming significant inhibition of the progression of brain infection (
P<0.05).
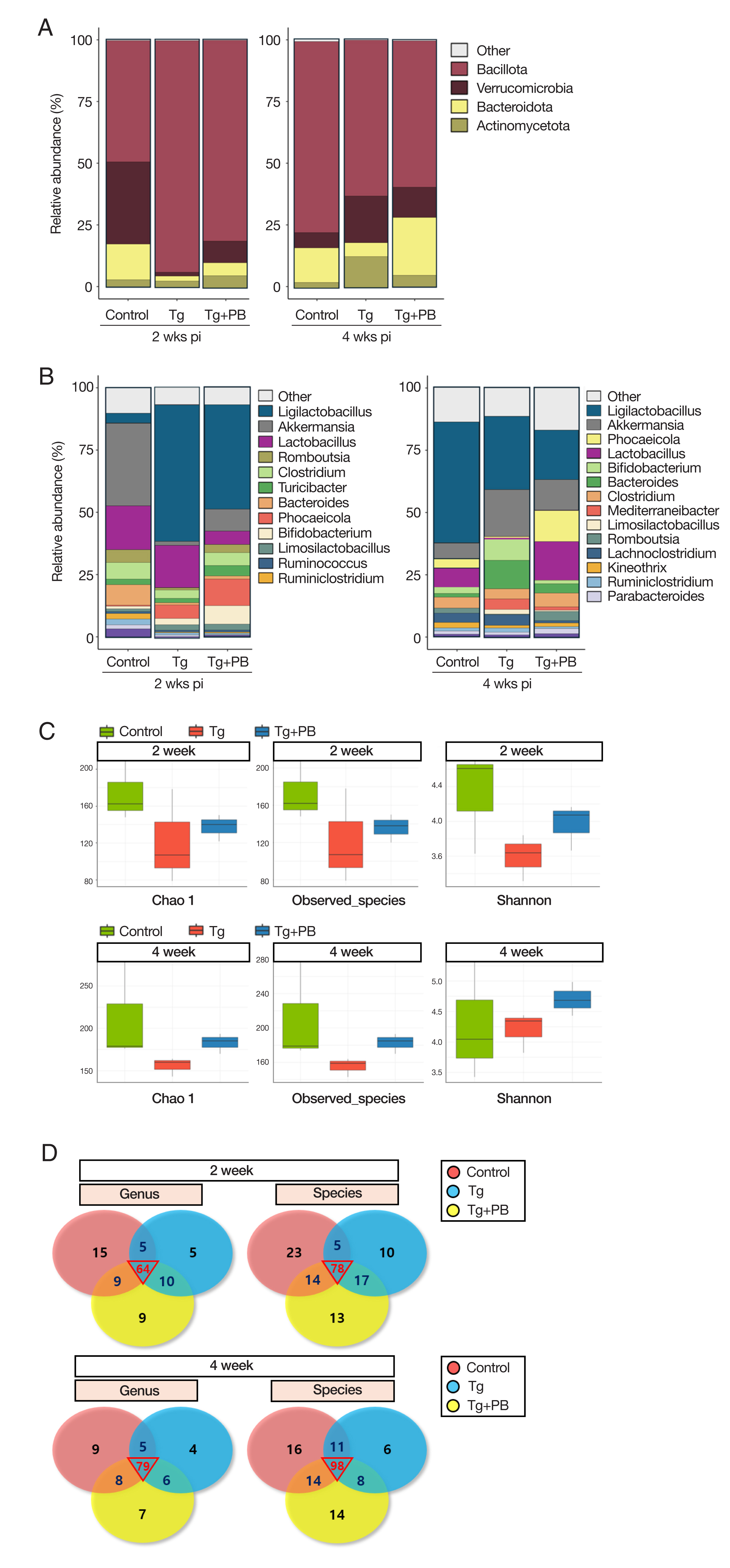
We used the ASV abundance and taxonomy data to compare the microbiota between groups at different taxonomic levels (phylum, class, order, family, genus, and species). At the phylum level, the microbiota composition of the control group included
Bacillota (49.2%),
Verrucomicrobia (33.3%),
Bacteroidota (14.5%), and
Actinomycetota (2.8%) after 2 weeks of infection. After 4 weeks of infection, there was a noticeable increase in
Bacillota (76.8%) and decrease in
Verrucomicrobia (6.1%) (
Fig. 2A). At the genus level, after 2 weeks of infection, the control group was dominated by
Akkermansia (33.3%) and
Bacteroides (8.2%) compared with the 2 infection groups (Tg: 1.6% and 0.9% and TG+PB: 8.7% and 1.3%, respectively) (
Fig. 2B). After 4 weeks of infection, there was a dramatic decrease in the distribution of
Akkermansia (6.1%) and
Lactobacillus (2 weeks, 17.5%; 4 weeks, 3.8%) in favor of
Ligilactobacillus (2 weeks, 3.5%; 4 weeks, 48.7%) (
Fig. 2B). Conversely, the Tg group was dominated by
Bacillota (93.5%) at the phylum level after 2 weeks of infection, with
Verrucomicrobia (1.6%),
Bacteroidota (2.0%), and
Actinomyceta (2.4%) comprising <10% (
Fig. 2A). After 4 weeks of infection, the distribution of
Bacillota (62.8%) decreased and the proportion of
Verrucomicrobia (18.8%),
Bacteroidota (5.6%), and
Actinomyceta (12.4%) increased, each accounting for >30% (
Fig. 2A). At the genus level, the Tg group showed a decrease in
Akkermansia and
Bacteroides and an increase in
Ligilactobacillus (54.8%) compared with the control group after 2 weeks of infection (
Fig. 2B). After 4 weeks of infection, the Tg group showed a decreased composition of
Ligilactobacillus (29.4%) and
Lactobacillus (2 weeks, 17.0%; 4 weeks, 0.6%) and an increased composition of
Clostridium (2 weeks, 3.4%; 4 weeks, 8.2%) and
Bifidobacterium (2 weeks, 1.7%; 4 weeks, 11.4%) (
Fig. 2B). At the phylum level (
Fig. 2A), the Tg+PB group showed recovered levels of
Verrucommicrobia,
Bacteroidota, and
Actinomycetota, which were reduced after 2 weeks of infection in the Tg group compared with the control group. After 4 weeks of infection, the share of the total decreased by approximately 20% for
Bacillota, with
Verrucomicrobia and
Bacteroidota taking its place and
Bacteroidota increasing from 5.2% to 23.4% (
Fig. 2A). At the genus level (
Fig. 2B), after 2 weeks of infection, the Tg+PB group showed a higher abundance of
Romboutsia (10.7%) and
Turicibacter (7.4%) compared with the other 2 groups (control: 0.4% and 1.1% and Tg: 0.9% 2.5%, respectively). Unexpectedly, the distribution of
Ligilactobacillus (41.8%) and
Lactobacillus (5.4%) in the Tg+PB group was lower than that in the Tg group (54.8% and 17.0%, respectively) (
Fig. 2B). After 4 weeks of infection,
Lactobacillus comprised the highest proportion in all 3 groups, and the abundance of “other,” a group comprising taxa with <1% abundance, increased to 16.9% in the Tg+PB group, which was slightly higher than the other 2 groups (control: 13.9% and Tg: 11.4%) (
Fig. 2B).
We analyzed diversity by assessing the species richness and evenness of gut microbes in addition to their numbers. To identify changes in the shared microbiota between groups, we analyzed and compared the observed species. Chao1 and Shannon indices in fecal samples from each group and visualized the taxonomic assignments of each group were using a Venn diagram. After 2 weeks of infection, the Chao1 and Shannon indices in the Tg and Tg+PB groups were all decreased compared with the control. However, the Shannon index in the Tg+PB group was increased (
Fig. 2C). After 4 weeks of infection, the Tg+PB group had higher results for all indices compared with the Tg group (
Fig. 2C). The Venn diagram revealed that the number of microbial species gradually increased in all groups over time. Furthermore, the number of ASVs shared between groups was higher after 4 weeks of infection compared with after 2 weeks of infection (
Fig. 2D).
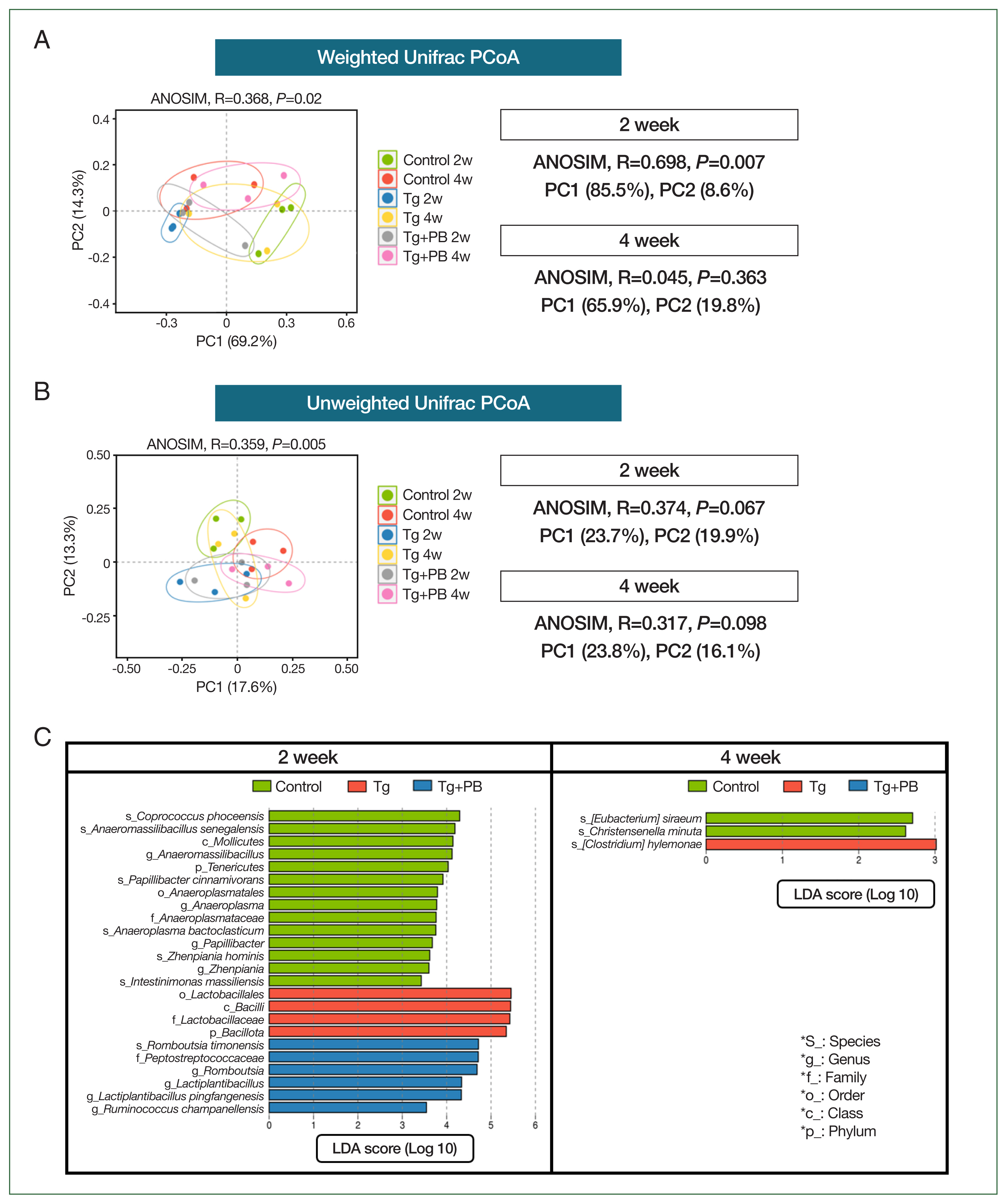
We used multivariate statistical analyses based on weighted and unweighted UniFrac distances to analyze the gut microbiota in the control (Tg and Tg+PB groups).
Fig. 3A presents the gut microbiota composition of each group according to PCoA based on weighted UniFrac distance, which considers not only the presence of microbes but also their relative abundance. The ANOSIM
R-value for the dataset of all groups after 2 and 4 weeks of infection was 0.368 (
P=0.02). This indicated that the Tg+PB groups after 2 and 4 weeks of infection were significantly differentiated from each other. Thus, the gut microbiota composition of the Tg+PB groups was somewhat distinguishable, with similarities between samples within the groups and lower similarities between the other groups. The most striking result was the very high
R-value (0.698,
P<0.007) after 2 weeks of infection, indicating a very low similarity between the 3 groups (control, Tg, and Tg+PB). However, after 4 weeks of infection, the results between the groups were indistinguishable (
Fig. 3A). We then performed PCoA based on unweighted UniFrac distance, which considers only differences in the presence or absence of gut microbiota. The results appeared similar to those of the PCoA based on weighted UniFrac distance (
R-value=0.359,
P=0.005) (
Fig. 3B). However, the differences in gut microbiota between groups were unclear and not statistically significant after both 2 and 4 weeks of infection. LEfSe analysis was then performed to further examine the differences in gut microbiota between the control, Tg, and Tg+PB groups after 2 and 4 weeks of infection (
Fig. 3C). The number of data (taxons) with a significant difference between the 3 groups (control, Tg, and Tg+PB) was higher after 2 weeks than after 4 weeks of infection (
P<0.05,
Fig. 3C).
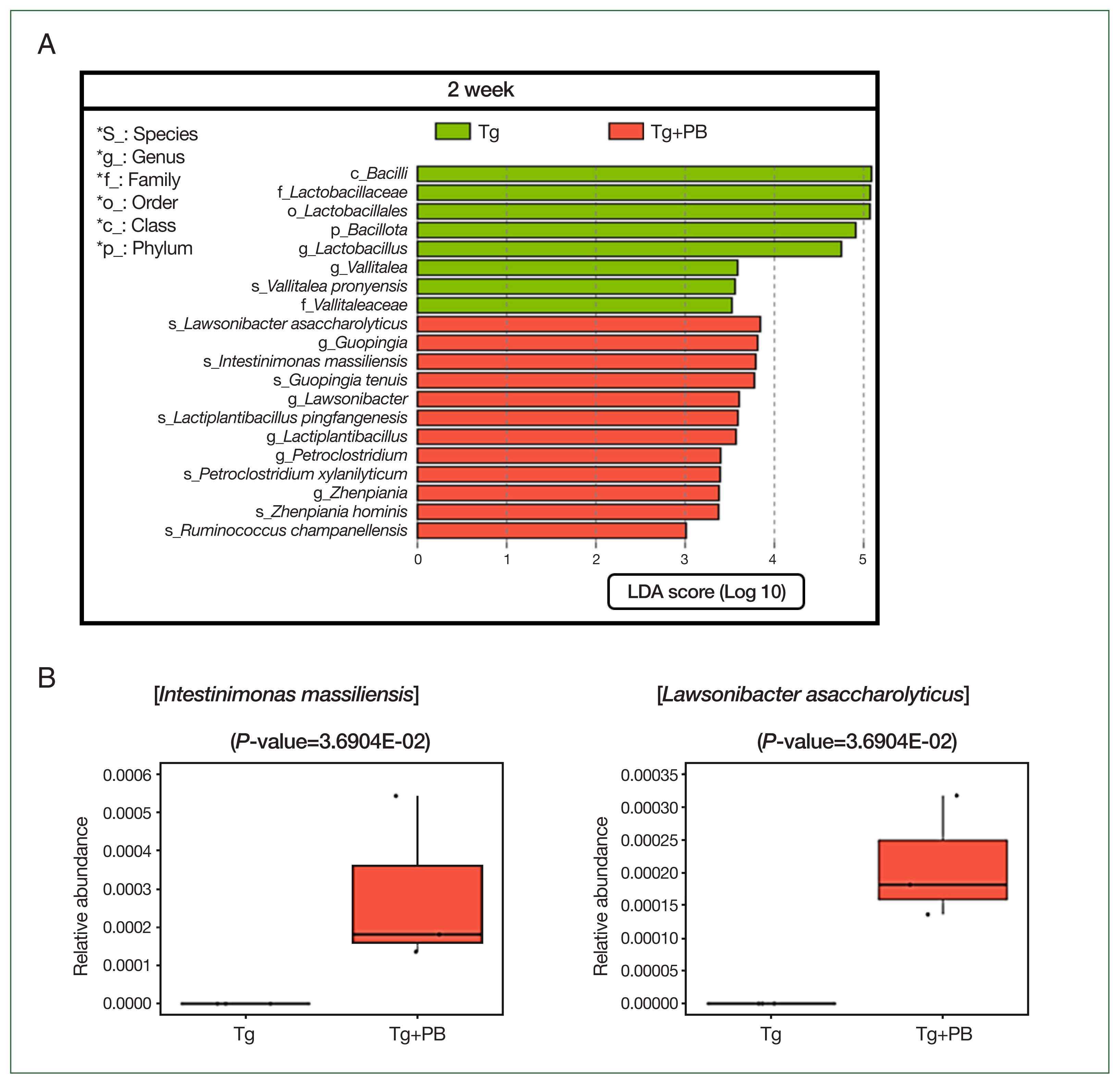
As observed in our previous analyses, there were notable disparities between the treatment groups after 2 weeks of infection. Thus, we aimed to pinpoint microbial species exhibiting statistically significant differentiations between the Tg and Tg+PB groups after 2 weeks of infection to elucidate potential microbial influencers of
T. gondii infection. As in our previous analysis, we used the LEfSe method to identify microbial species with significant differences in abundance between the Tg and Tg+PB groups.
Vallitalea pronyensis was the only microbial species that was decreased in the Tg+PB group compared with the Tg group after 2 weeks of infection (
Fig. 4A). In contrast, seven microbial species were increased compared with the Tg group after 2 weeks of infection:
Lactiplantibacillus pingfangensis,
Guopingia tenuis,
Zhenpiania hominis,
Ruminococcus champanellensis,
Petroclostridium xylanilyticum,
Intestinimonas massiliensis, and
Lawsonibacter asaccharolyticus (
Fig. 4A). Among them,
I. massiliensis and
Lawsonibacter asaccharolyticus, known as involved in SCFA production [
19], were significantly increased in the Tg+PB group compared with the Tg group after 2 weeks of infection (
P<0.05) (
Fig. 4B).
We hypothesized that changes in the gut microbiota caused by probiotic supplementation and the resultant functional changes in the gut would reduce
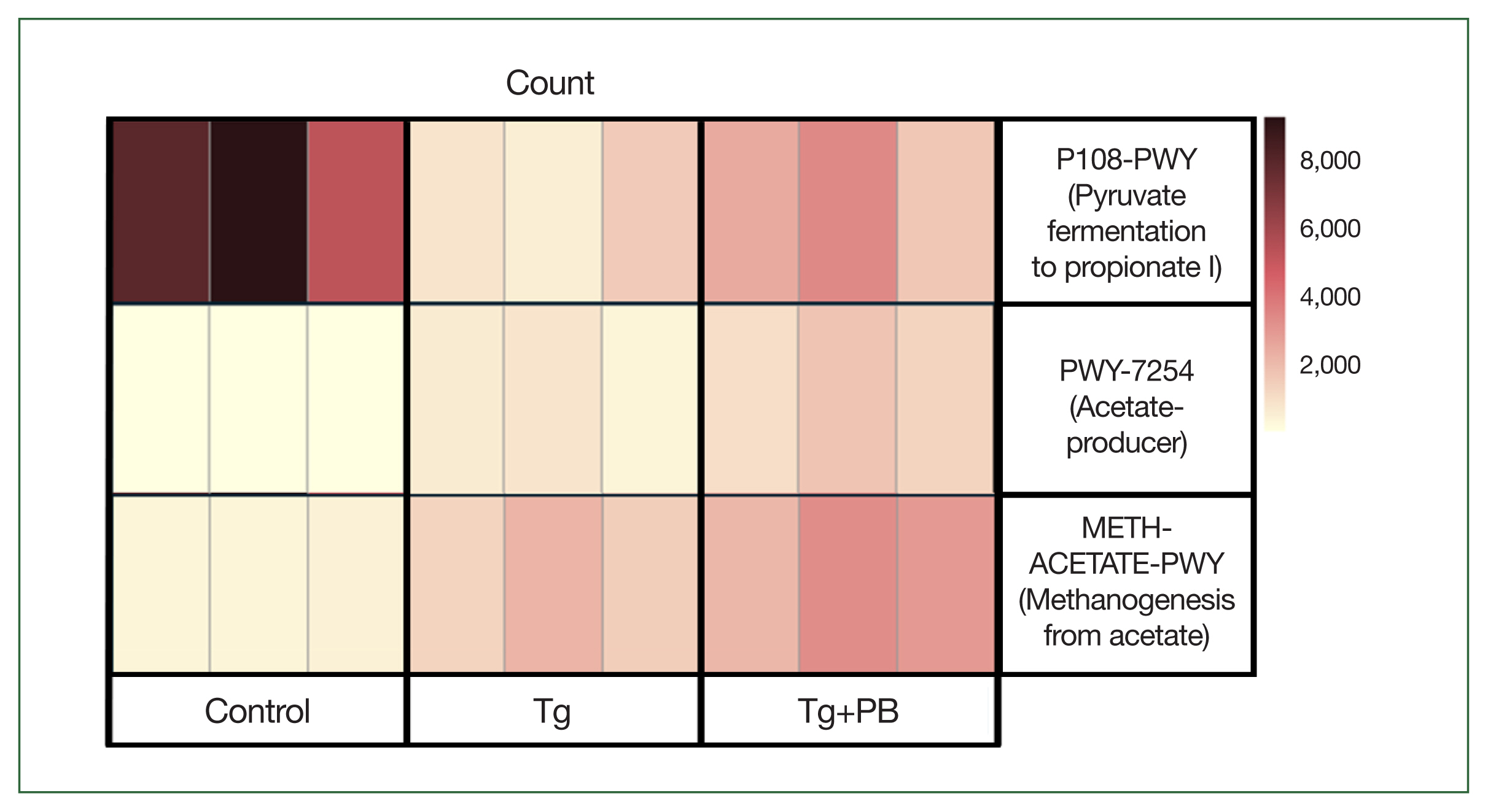
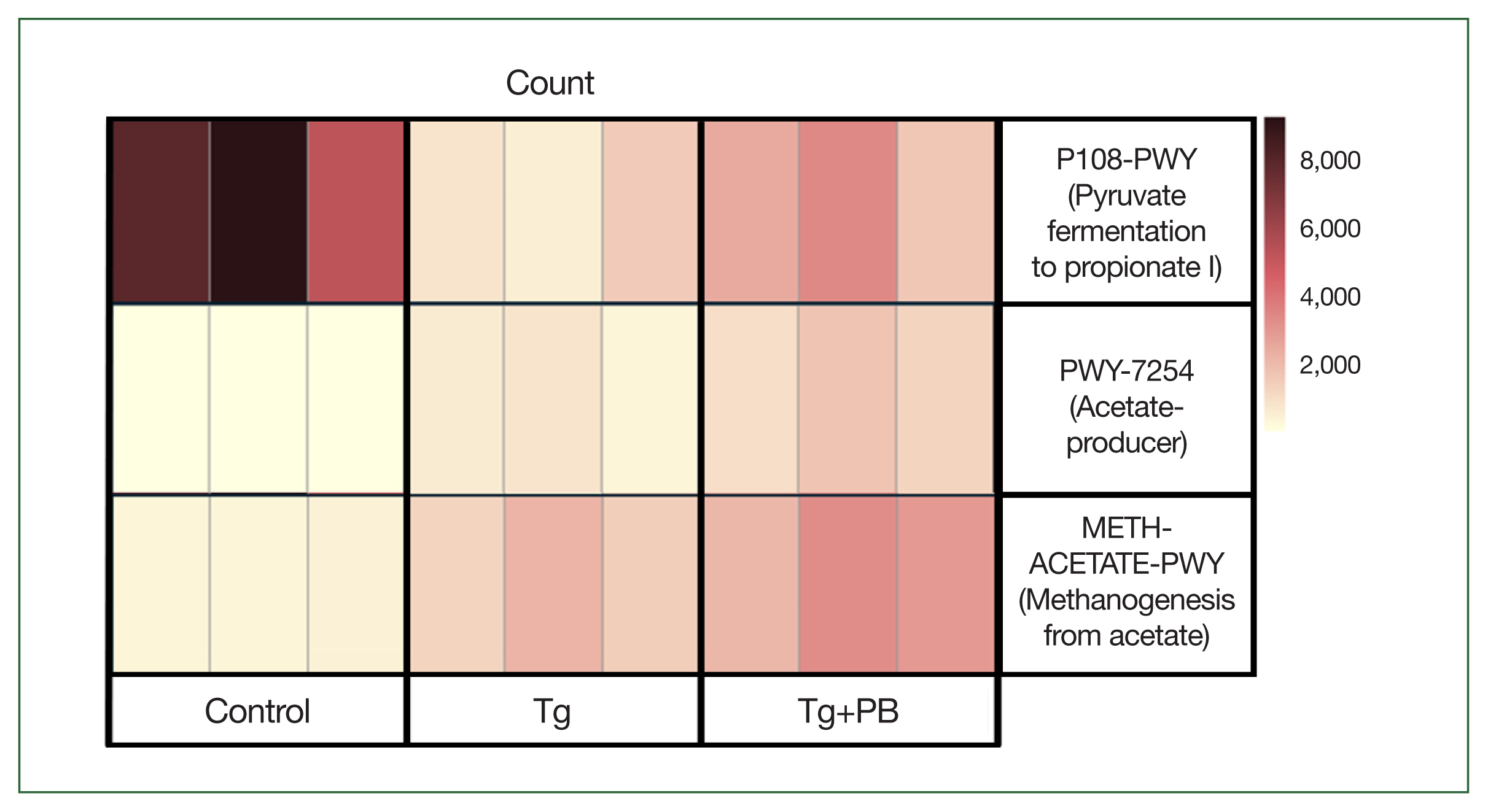
T. gondii levels in the brain of infected mice. Our previous analyses revealed an increase in bacterial species involved in SCFA production. To further investigate this, we scored functional changes in samples from each group using PICRUSt2 analysis to determine whether the abundance scores of pathways involved in SCFA production were significantly higher in the Tg+PB group compared with the Tg group. METH-ACETATE-PWY (acetate to methane pathway), PWY-7254 (TCA cycle VII acetate producers pathway), and P108-PWY (pyruvate fermentation to propionate I pathway) were upregulated in the Tg+PB group (
Fig. 5). Notably, among these pathways, P108-PWY exhibited a marked reduction in the Tg group compared with the control group.
Discussion
Toxoplasma gondii infects all warm-blooded animals, including humans, and is highly prevalent worldwide [
5]. Infection occurs through the ingestion of cysts or oocysts in undercooked meat or food and the contaminated water. Following infection,
T. gondii spreads to various organs until it transitions from the rapidly dividing tachyzoite stage to the latent bradyzoite stage in response to the host’s immune response.
T. gondii initially infects the host’s intestinal epithelial cells, proliferates within the infected cells, and then exits them to infect neighboring cells [
3]. Thus, proliferating
T. gondii can be observed in the intestinal epithelium in the early stages of infection [
20]. After entering the bloodstream through the intestinal epithelium,
T. gondii rapidly reaches the brain and produces cysts. Brain cysts can be detected as early as 2 weeks after infection, increasing in number for up to 4 weeks and then reaching a plateau [
21]. Mice orally infected with
T. gondii for 7 days were reported to show an increase in persistent
T. gondii infection in the ileum parenchyma and Peyer’s patches [
22]. Furthermore, it is well known that changes in the intestinal microbial composition affect the intestinal mucosa and immune function [
11]. The gut microbiota stimulates dendritic cells in a TLR11-independent manner to induce a response from Th1 cells to
T. gondii infection [
23]. The role of the gut microbiota in infection is an active area of research. A study on changes in the gut microbiota composition of mice infected with
T. gondii and fed fermented dairy products revealed that the number of brain cysts in the fermented dairy group was reduced compared with the control group [
24]. Our study used next-generation sequencing techniques to investigate the impact of probiotic supplementation on the gut microbiota of
T. gondii-infected mice and its potential to mitigate
T. gondii infection.
We used qPCR to compare the levels of
T. gondii-specific B1 gene DNA levels to determine the inhibitory effects of probiotic supplementation on
T. gondii infection (
Fig. 1). Brain cysts can be detected after 2 weeks of infection, peaking after 4 weeks and then plateauing [
21]. Thus, we investigated the effect of probiotic supplementation on
T. gondii infection by harvesting the brains of infected mice after 2 and 4 weeks of infection to compare B1 gene DNA levels between groups. We also investigated gut microbiota composition and functional changes during this time.
An increased proportion of
Bacillota in
T. gondii-infected intestine may induce an inflammatory environment in the gut, thereby compromising the integrity and permeability of the intestinal epithelial barrier. This altered environment potentially enhances
T. gondii penetration through the intestinal wall, promoting migration into deeper tissues. We observed a decline in
Bacillota and an upsurge in
Verrucomicrobia,
Bacteroidota, and
Actinomycetota in the Tg+PB group compared with the Tg group. The phylum
Verrucomicrobia includes
Akkermansia muciniphila, renowned for its anti-inflammatory properties and role in intestinal wound healing [
25].
Bifidobacterium belongs to the phylum
Actinomycetota, and its augmentation may have contributed to promoting intestinal health [
15]. Notably, at the genus level, there was an increase in
Romboutsia and
Phocaeicola in the Tg+PB group compared with the Tg group after 2 weeks of infection.
Romboutsia is abundant in healthy mucous membranes and
Phocaeicola produces SCFAs that help to heal the gut wall [
26]. Consequently, this alteration may strengthen the intestinal wall, playing a role in mitigating
T. gondii infection in the gut. Interestingly,
Lactobacillus and
Ligilactobacillus exhibited increased abundance in the Tg group relative to the control group at week 2. Although augmented
Lactobacillus levels have also been reported in studies on parasites,
Helicobacter pylori infections, and diverse diseases, the underlying cause and function of this increase remains unclear [
19]. By week 4, the Tg group exhibited an elevation in
Bacteroides and
Akkermansia at the genus level. This phenomenon is presumably attributed to the migration of most
T. gondii parasites from the intestine to other organs, resulting in decreased infectivity and subsequent restoration of intestinal microbiota diversity and stability. The Tg+PB group also appeared to stabilize with increased bacterial richness. The higher Shannon index in the Tg+PB group compared with the Tg group suggests that probiotic supplementation improved the species diversity reduced by
T. gondii infection, supported by the increased Shannon index in the Tg+PB group at weeks 2 and 4 compared with the Tg group. This observation suggests that probiotics positively impact gut microbial diversity. At week 4, there was a trend toward a more similar gut environment, with an increase in the number of taxa shared between groups compared with week 2, likely due to microbiota stabilization after a sharp decline in
T. gondii infection in the gut.
The timing of
T. gondii alteration of the gut microbiota was explored using PCoA plots and comparing gut microbiota between groups. It has been confirmed that changes in the gut microbiota are strongly driven by the acute phase of
T. gondii infection [
27]. This supports our findings of a clear separation between the control, Tg, and Tg+PB groups at week 2. However, by week 4, the differences between groups became indistinct. The PCoA plots based on unweighted UniFrac distance showed less difference in gut microbiota between groups than the PCoA results based on weighted UniFrac distance. Furthermore, the subsequent LEfSe analysis confirmed that the difference in gut microbiota composition between groups was reduced at week 4 compared with week 2. This suggests that following
T. gondii infection, the distinction in gut microbiota composition between groups decreased over time. We compared brain infection levels using qPCR, revealing increased infection in the Tg+PB group compared with the Tg group at week 2. However, by week 4, the infection level in the Tg+PB group was reduced compared with the Tg group. Thus, our findings indicate that following intestinal infection,
T. gondii requires approximately 2 weeks to migrate to the brain and undergo cyst formation. Thereafter, intestinal penetration by
T. gondii diminishes. Consequently, the significant reduction in brain infection observed in the Tg+PB group at week 4 can be attributed to the influence of the gut microbiota at week 2 (
P<0.05). This is supported by our qPCR results and is consistent with the distinct separation between groups in the PCoA plots at week 2.
We observed a increase in
I. massiliensis and
Lawsonibacter asaccharolyticus in the Tg+PB group compared with the Tg group. These bacteria are involved in SCFA production, such as butyrate, acetate, and propionate [
28]. SCFAs stimulate intestinal mucus production, which protects the intestinal epithelium, strengthens tight junctions, and reduces inflammation [
29]. Propionate reduces intestinal inflammation and protects intestinal wall function by increasing tight junction proteins [
30]. Butyrate exerts anti-inflammatory effects by inhibiting NF-κB and IFN-γ and upregulating PPAR-γ [
31]. The qPCR results at week 2 showed a significant increase in infection levels in the Tg+PB group (
P<0.05), possibly due to the creation of a more favorable environment for
T. gondii proliferation in early infection.
T. gondii is generally present in the brain after approximately 1 week of infection [
32]. In other words, some
T. gondii have already invaded the brain in the second week. Thus, this period can be considered unaffected by probiotic supplementation. Furthermore, because the difference in B1 gene expression between Tg and Tg+PB groups was not significant at week 2, It was suspected that differences in the timing of the interventions might affect B1 gene expression levels. The role of butyrate, an SCFA, in strengthening and repairing the barrier function of intestinal epithelial cells has been established [
33]. Thus, the anti-inflammatory response may heal and strengthen the damaged gut wall over time, thereby preventing further
T. gondii invasion and reducing infection levels at week 4, as reflected in our qPCR results. However, further studies are necessary to fully elucidate this complex interaction.
We performed PICRUSt2 analysis to predict the functional changes in the gut microbiota between the groups at week 2, a time point at which distinct alterations in the gut microbiota were observed due to
T. gondii infection and probiotic supplementation. It has been suggested that
T. gondii infection significantly impacts the metabolic function of the gut microbiota [
27]. Furthermore, SCFAs reduce inflammation, thicken the mucosal layer, improve intestinal wall function, and prevent gut damage [
34]. Based on these data and the previous findings [
28], we predicted that probiotics would increase the activity of functional pathways involved in SCFA production in the Tg+PB group. We initially focused on whether there was a statistically significant increase in the activity of SCFA-related functional pathways in the Tg+PB group compared with the Tg group. This led to the discovery of increased P108-PWY, METH-ACETATE-PWY, and PWY-7254 activity in the Tg+PB group compared with the Tg group. METH-ACETATE-PWY activation suggests enhanced methanogenesis from acetate. However, this increase may also be due to increased acetate production [
35]. Considering the simultaneous upregulation of PWY-7254, which is involved in acetate production, it is plausible that the observed increase in METH-ACETATE-PWY activity is a response to an intestinal environment characterized by increased acetate availability. In the Tg+PB group, we expected a functional elevation in a pathway that was involved in the production of propionate, a major SCFA that may contribute to improved gut barrier function [
30]. This expectation was supported by the upregulation of P108-PWY. P108-PWY activity was reduced in the Tg group compared with the control group, and the deterioration of this functional pathway was mitigated by probiotics. This suggests an increase in the intestinal content of propionate, thereby enhancing barrier function to suppress
T. gondii infection by limiting access to new infections and preventing deeper penetration into tissues. Thus, probiotic-induced modification of this functional pathway may help to suppress
T. gondii infection.
In conclusion, our study suggests that probiotic-induced changes in the gut microbiota and modulation of gut microbial function to suppress T. gondii infection. Alterations in the gut environment induced by probiotic supplementation during T. gondii infection likely enhance SCFA production. These effects collectively contribute to mitigating T. gondii infection severity. However, a limitation of our study is the focus on changes in the fecal microbiota without investigating the detailed biological and mechanistic contributions of SCFAs to inhibiting T. gondii infection. Further in-depth studies are warranted to determine whether the observed increase in SCFAs directly contributed to infection suppression.
Notes
-
Author contributions
Conceptualization: Lee HJ, Shin EH
Data curation: Lee HJ, Ham DW, Shin EH
Formal analysis: Lee HJ, Ham DW
Funding acquisition: Shin EH
Investigation: Lee HJ, Ham DW, Seo SH, Cha GH
Methodology: Seo SH
Project administration: Shin EH
Resources: Cha GH, Shin EH
Supervision: Shin EH
Writing – original draft: Lee HJ
Writing – review & editing: Shin EH
-
The authors declare no conflict of interest related to this study.
Fig. 1Level of B1 gene DNA in Toxoplasma gondii-infected mice. The T. gondii-specific B1 gene was used as a marker to assess the extent of T. gondii infection. Bars represent fold changes in B1 gene DNA compared with the control. Control, uninfected group; Tg, T. gondii-infected group; Tg+PB, T. gondii-infected group receiving probiotic supplementation. *P<0.05 compared with the control group, †P<0.05 between groups.
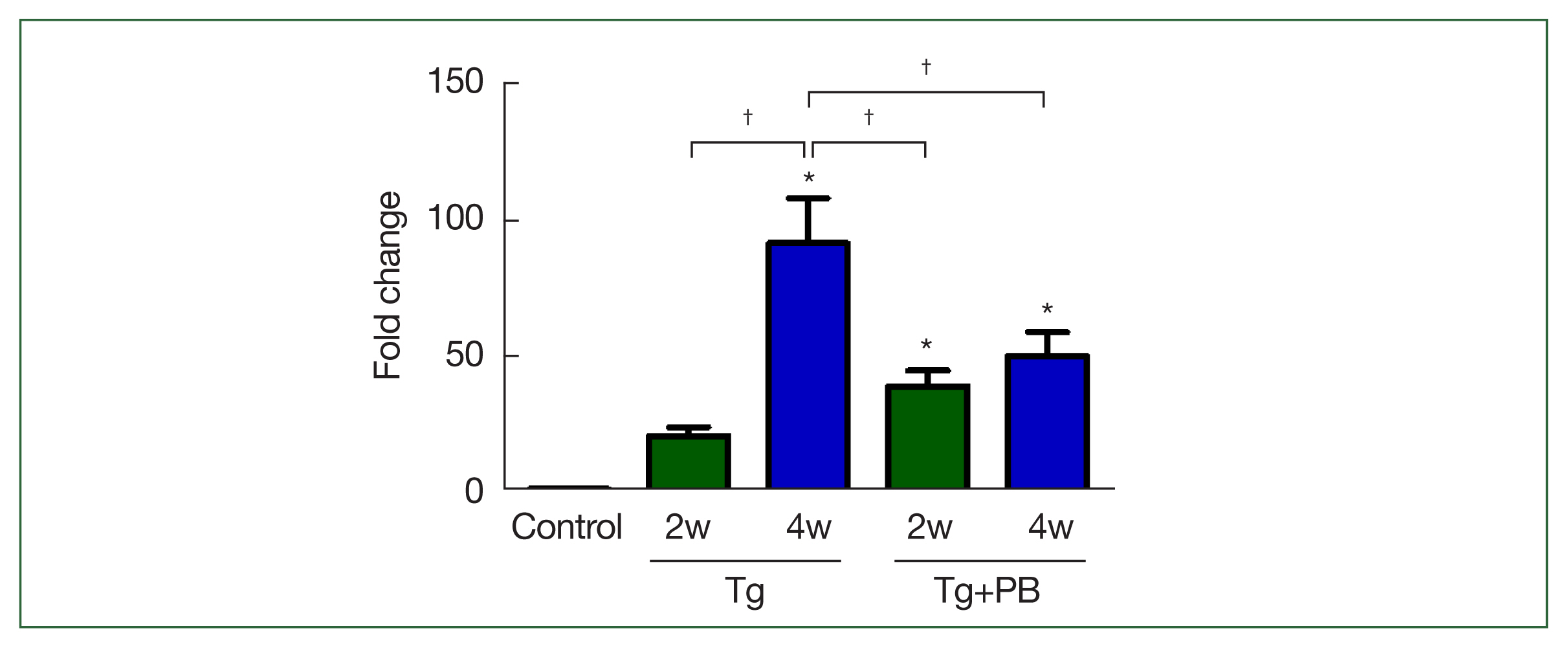
Fig. 2Comprehensive analysis of microbial community composition and diversity across taxonomic levels. (A) Bar graph of the relative abundance of microbial communities at the phylum level. (B) Bar graph of the microbial community composition at the genus level. (C) Microbial diversity analysis of fecal samples. The observed species, Chao1, and Shannon indices reflect species abundance. richness, and evenness, respectively. (D) Venn diagrams of shared and unique microbial taxa at the genus and species levels in fecal samples. Numbers in each section of the circle represent the number of taxa shared or unique to the group.
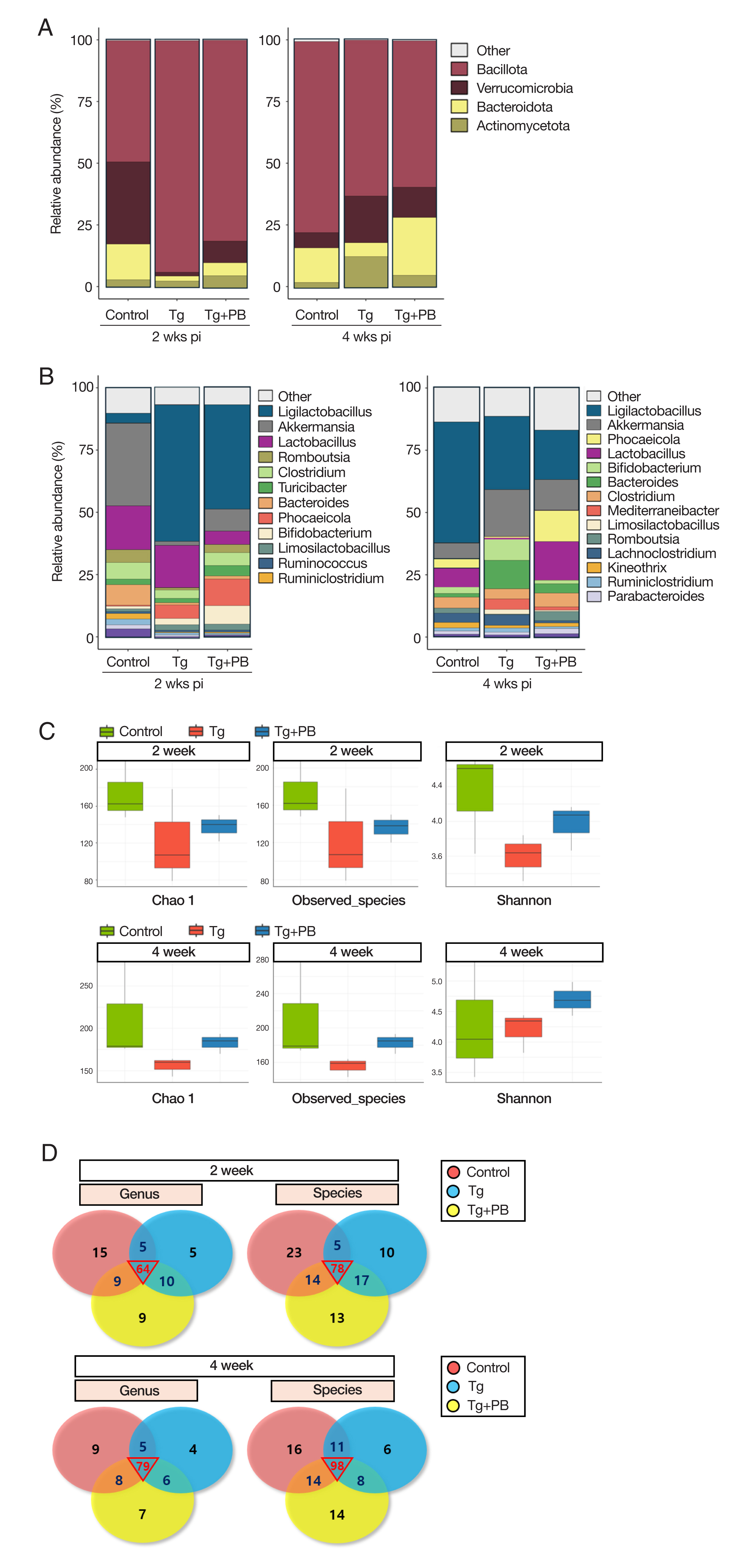
Fig. 3Comparative analysis of microbial community changes. (A) PCoA results of microbial communities based on weighted UniFrac distances across different treatment groups and time points. ANOSIM was used to determine the statistical significance of differences between groups. (B) PCoA results of microbial communities based on unweighted UniFrac distances (C). Graph presenting the results of a LEfSe analysis performed to identify statistically significant differences in the microbial taxa across the control, Tg, and Tg+PB groups after 2 and 4 weeks of Toxoplasma gondii infection. Each bar represents a significantly enriched microbial taxon in one of the groups. Bar length indicates the extent of the difference.
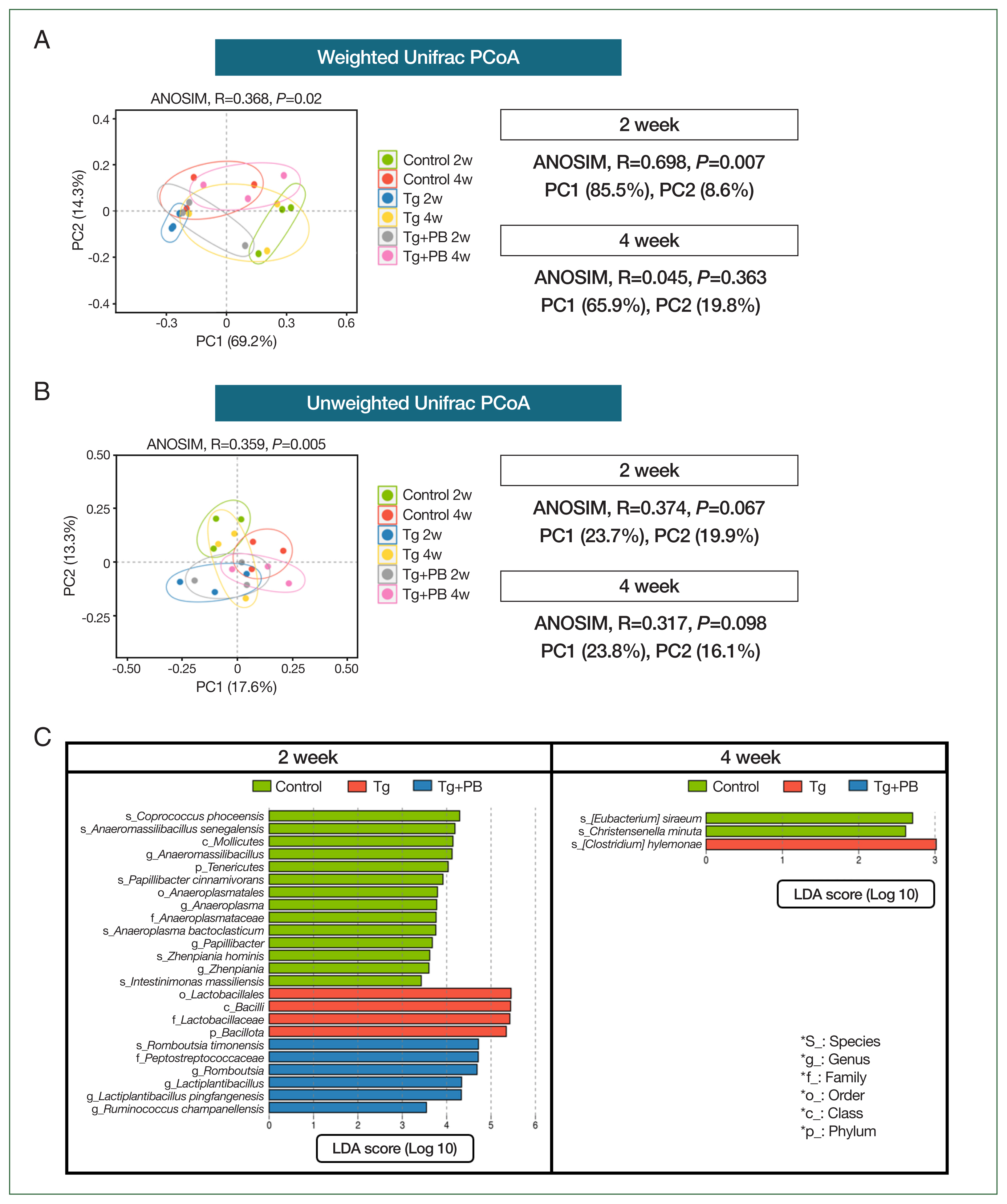
Fig. 4LEfSe analysis of microbial differences between the Tg and Tg + PB groups after 2 weeks of infection with Toxoplasma gondii. (A) Bar graph showing the results of the LEfSe analysis performed to determine significant differences in microbial species between the Tg and Tg+PB groups after 2 weeks of infection with Toxoplasma gondii. (B) Boxplot showing the substantial increase in Intestinimonas massiliensis and Lawsonibacter asaccharolyticus in the Tg+PB group after 2 weeks of infection.
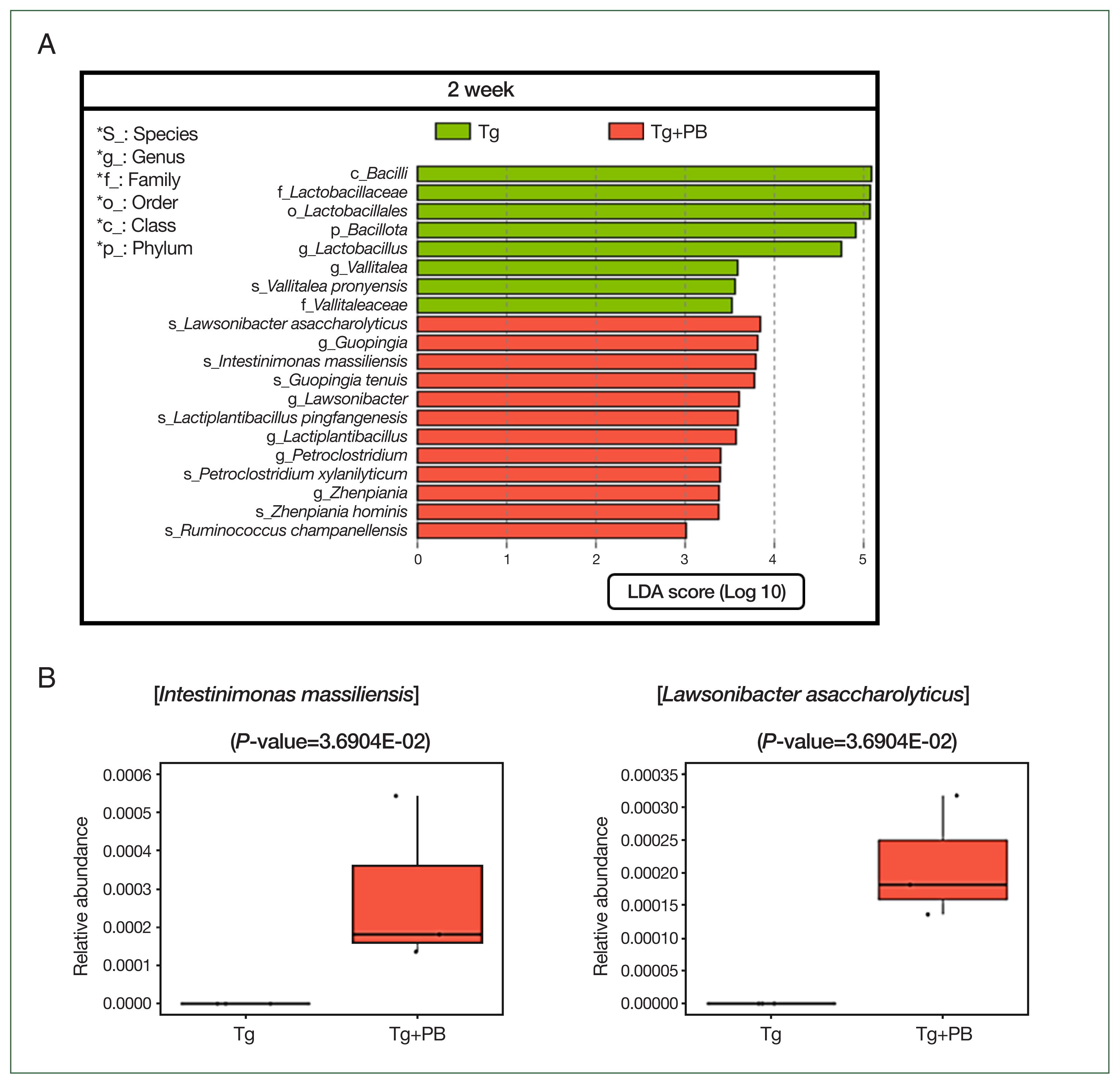
Fig. 5Heatmap of pathway abundance scores in gut microbiota influenced by probiotic supplementation during Toxoplasma gondii infection. The heatmap is a visual representation of functional changes in the gut microbiota induced by probiotic supplementation after 2 weeks of T. gondii infection in mice. Pathway abundance scores derived from PICRUSt2 analysis highlight critical functional changes in microbial metabolism.
References
- 1. Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, et al. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 2011;17(1):7.
https://doi.org/10.3201/eid1701.P11101
- 2. Kortbeek LM, Hofhuis A, Nijhuis CDM, Havelaar AH. Congenital toxoplasmosis and DALYs in the Netherlands. Mem Inst Oswaldo Cruz 2009;104(2):370-373.
https://doi.org/10.1590/S0074-02762009000200034
- 3. Blader IJ, Coleman BI, Chen CT, Gubbels MJ. Lytic cycle of Toxoplasma gondii: 15 years later. Annu Rev Microbiol 2015;69:463-485.
https://doi.org/10.1146/annurev-micro-091014-104100
- 4. Feustel SM, Meissner M, Liesenfeld O.
Toxoplasma gondii and the blood-brain barrier. Virulence 2012;3(2):182-192.
https://doi.org/10.4161/viru.19004
- 5. Matta SK, Rinkenberger N, Dunay IR, Sibley LD.
Toxoplasma gondii infection and its implications within the central nervous system. Nat Rev Microbiol 2021;19(7):467-480.
https://doi.org/10.1038/s41579-021-00518-7
- 6. Atmaca LS, Simsek T, Batioglu F. Clinical features and prognosis in ocular toxoplasmosis. Jpn J Ophthalmol 2004;48:386-391.
https://doi.org/10.1007/s10384-003-0069-0
- 7. Sutterland AL, Fond G, Kuin A, Koeter MWJ, Lutter R, et al. Beyond the associationToxoplasma gondii in schizophrenia, bipolar disorder, and addiction: systematic review and meta-analysis. Acta Psychiatr Scand 2015;132(3):161-179.
https://doi.org/10.1111/acps.12423
- 8. Dunay IR, Gajurel K, Dhakal R, Liesenfeld O, Montoya JG. Treatment of toxoplasmosis: historical perspective, animal models, and current clinical practice. Clin Microbiol Rev 2018;31(4):10-1128.
https://doi.org/10.1128/cmr.00057-17
- 9. Snyder LM, Denkers EY. From initiators to effectors: roadmap through the intestine during encounter of Toxoplasma gondii with the mucosal immune system. Front Cell Infect Microbiol 2021;10:614701.
https://doi.org/10.3389/fcimb.2020.614701
- 10. Partida-Rodríguez O, Serrano-Vázquez A, Nieves-Ramírez ME, Moran P, Rojas L, et al. Human intestinal microbiota: interaction between parasites and the host immune response. Arch Med Res 2017;48(8):690-700.
https://doi.org/10.1016/j.arcmed.2017.11.015
- 11. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell 2014;157(1):121-141.
https://doi.org/10.1016/j.cell.2014.03.011
- 12. Yan X, Sun Y, Zhang G, Han W, Gao J, et al. Study on the antagonistic effects of koumiss on Toxoplasma gondii infection in mice. Front Nutr 2022;9:1014344.
https://doi.org/10.3389/fnut.2022.1014344
- 13. Kumar Bajaj B, Claes IJ, Lebeer S. Functional mechanisms of probiotics. J Microbiol Biotechnol Food Sci 2015;4(4):321-327.
https://doi.org/10.15414/JMBFS.2015.5.2.109-115
- 14. Libertucci J, Young VB. The role of the microbiota in infectious diseases. Nat Microbiol 2019;4(1):35-45.
https://doi.org/10.1038/s41564-018-0278-4
- 15. Hiippala K, Jouhten H, Ronkainen A, Hartikainen A, Kainulainen V, et al. The potential of gut commensals in reinforcing intestinal barrier function and alleviating inflammation. Nutrients 2018;10(8):988.
https://doi.org/10.3390/nu10080988
- 16. Zhang Z, Lv J, Pan L, Zhang Y. Roles and applications of probiotic Lactobacillus strains. Appl Microbiol Biotechnol 2018;102:8135-8143.
https://doi.org/10.1007/s00253-018-9217-9
- 17. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, et al. Metagenomic biomarker discovery and explanation. Genome Biol 2011;12(6):
https://doi.org/10.1186/gb-2011-12-6-r60
- 18. Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol 2020;38(6):685-688.
https://doi.org/10.1038/s41587-020-0548-6
- 19. Rossi F, Amadoro C, Colavita G. Members of the Lactobacillus genus complex (LGC) as opportunistic pathogens: a review. Microorganisms 2019;7(5):126.
https://doi.org/10.3390/microorganisms7050126
- 20. Speer CA, Dubey JP. Ultrastructure of early stages of infections in mice fed Toxoplasma gondii oocysts. Parasitology 1998;116(1):35-42.
https://doi.org/10.1017/S0031182097001959
- 21. Chew WK, Wah MJ, Ambu S, Segarra I.
Toxoplasma gondii: determination of the onset of chronic infection in mice and the in vitro reactivation of brain cysts. Exp Parasitol 2012;130(1):22-25.
https://doi.org/10.1016/j.exppara.2011.10.004
- 22. Mitsunaga T, Norose K, Aosai F, Horie H, Ohnuma N, et al. Infection dynamics of Toxoplasma gondii in gut-associated tissues after oral infection: the role of Peyer’s patches. Parasitol Int 2019;68(1):40-47.
https://doi.org/10.1016/j.parint.2018.08.010
- 23. Benson A, Pifer R, Behrendt CL, Hooper LV, Yarovinsky F. Gut commensal bacteria direct a protective immune response against Toxoplasma gondii. Cell Host Microbe 2009;6(2):187-196.
https://doi.org/10.1016/j.chom.2009.06.005
- 24. Dubey JP. Bradyzoite-induced murine toxoplasmosis: stage conversion, pathogenesis, and tissue cyst formation in mice fed bradvzoites of-different strains of Toxoplasma gondii. J Eukaryot Microbiol 1997;44(6):592-602.
https://doi.org/10.1111/j.1550-7408.1997.tb05965.x
- 25. Wade H, Pan K, Duan Q, Kaluzny S, Pandey E, et al.
Akkermansia muciniphila and its membrane protein ameliorates intestinal inflammatory stress and promotes epithelial wound healing via CREBH and miR-143/145. J Biomed Sci 2023;30(1):38.
https://doi.org/10.1186/s12929-023-00935-1
- 26. Lück R, Deppenmeier U. Genetic tools for the redirection of the central carbon flow towards the production of lactate in the human gut bacterium Phocaeicola (Bacteroides) vulgatus. Appl Microbiol Biotechnol 2022;106(3):1211-1225.
https://doi.org/10.1007/s00253-022-11777-6
- 27. Meng JX, Wei XY, Guo H, Chen Y, Wang W, et al. Metagenomic insights into the composition and function of the gut microbiota of mice infected with Toxoplasma gondii. Front Immunol 2023;14:1156397.
- 28. Afouda P, Durand GA, Lagier JC, Labas N, Cadoret F, et al. Noncontiguous finished genome sequence and description of Intestinimonas massiliensis sp. nov strain GD 2T, the second Intestinimonas species cultured from the human gut. Microbiologyopen 2019;8(1):e00621.
https://doi.org/10.1002/mbo3.621
- 29. Markowiak-Kopeć P, Śliżewska K. The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutrients 2020;12(4):1107.
https://doi.org/10.3390/nu12041107
- 30. Tong LC, Wang Y, Wang ZB, Liu WY, Sun S, et al. Propionate ameliorates dextran sodium sulfate-induced colitis by improving intestinal barrier function and reducing inflammation and oxidative stress. Front Pharmacol 2016;7:253.
https://doi.org/10.3389/fphar.2016.00253
- 31. Jiang L, Wang J, Liu Z, Jiang A, Li S, et al. Sodium butyrate alleviates lipopolysaccharide-induced inflammatory responses by down-regulation of NF-κB, NLRP3 signaling pathway, and activating histone acetylation in bovine macrophages. Front Vet Sci 2020;7:579674.
https://doi.org/10.3389/fvets.2020.579674
- 32. Mahamed DA, Mills JH, Egan CE, Denker EY, Bynoe MS. CD73-generated adenosine facilitates Toxoplasma gondii differentiation to long-lived tissue cysts in the central nervous system. Proc Natl Acad Sci USA 2012;109(40):16312-16317.
https://doi.org/10.1073/pnas.1205589109
- 33. Liang L, Liu L, Zhou W, Yang C, Mai G, et al. Gut microbiota-derived butyrate regulates gut mucus barrier repair by activating the macrophage/WNT/ERK signaling pathway. Clin Sci 2022;136(4):291-307.
https://doi.org/10.1042/CS20210778
- 34. Feng C, Jin C, Liu K, Yang Z. Microbiota-derived short chain fatty acids: their role and mechanisms in viral infections. Biomed Pharmacother 2023;160:114414.
https://doi.org/10.1016/j.biopha.2023.114414
- 35. Fotidis IA, Karakashev D, Kotsopoulos TA, Martzopoulos GG, Angelidaki I. Effect of ammonium and acetate on methanogenic pathway and methanogenic community composition. FEMS Microbiol Ecol 2013;83(1):38-48.
https://doi.org/10.1111/j.1574-6941.2012.01456.x