Performance validation of the BD MAX Enteric Parasite Panel using simulated samples in low endemic regions
Article information
Abstract
Molecular diagnostics are essential for detecting intestinal parasites, but evaluating clinical samples from low endemic areas, including Korea, is challenging. We tested the performance of the BD MAX Enteric Parasite Panel in simulated samples for clinical use. Simulated samples were prepared with residual stool samples to confirm the diagnostic performance of the kits. Standard materials for Cryptosporidium parvum, Giardia lamblia, and Entamoeba histolytica were obtained for assessment. Limit of detection was determined by diluting standard materials into multiple concentrations and testing each in duplicate. Repeatability was assessed by retesting all samples twice. Accuracy was evaluated by comparing BD MAX System results with intended results. The limit of detection values obtained using standard materials were 781 cysts/ml, 6,250 oocysts/ml, and 125 DNA copies/ml for G. lamblia, C. parvum, and E. histolytica, respectively. Simulated G. lamblia-positive stool samples with concentrations above 6,250 cysts/ml consistently yielded positive results (100% concordance). However, C. parvum-positive stool samples at 6,250 oocysts/ml showed 50% concordance initially and 75% after retesting. At 62,500 oocysts/ml, the concordance rates were 89% initially and 100% after retesting. Overall agreement was 95.2%, but that for C. parvum was relatively low (82.4%). The diagnostic performances were 87.8% of sensitivity and 100% of specificity. Despite the limited clinical samples, BD MAX Enteric Parasite Panel showed good performance for clinical use, and spiked samples proved useful for evaluating protozoan PCR in low-incidence regions.
Introduction
Enteric parasitic infections are important global health issue, necessitating prompt and accurate diagnosis for effective treatment and control [1]. Intestinal parasitic infections are a major cause of gastrointestinal diseases in developing countries, but they also cause illnesses in developed countries [2]. However, in low endemic countries, infections caused by intestinal parasites have been underestimated compared with those caused by other microorganisms because they are difficult to diagnose [3]. Traditional diagnostic methods such as microscopy are a major diagnostic tool in many laboratories, but they are labor intensive, require considerable knowledge and experience to interpret, and lack sensitivity and specificity for accurate diagnosis [4]. Molecular diagnostic tools, including multiplex PCR panels, are increasingly important for the diagnosis of intestinal parasites, with their utility as primary screening tools becoming well recognized [5]. Reliable commercial PCR assays are important to meet the increasing need for laboratory diagnosis. The BD MAX Enteric Parasite Panel (BD MAX EPP; BD Diagnostics, Baltimore, MD, USA) is a fully automated assay that provides nucleic acid extraction and simultaneous real-time amplification for the detection of Entamoeba histolytica, Giardia intestinalis, and Cryptosporidium parvum/hominis [6–9]. The test targets a Cryptosporidium-specific DNA fragment and small subunit rRNA genes for the other parasites [7]. This panel has been evaluated in multiple studies across different regions, demonstrating consistent performance in identifying protozoan infections [6–10]. Even though in vitro diagnostic assays are commercially available and Food and Drug Administration approved, each laboratory should verify whether these assays can reproduce the performance specifications in their laboratory for clinical use [11]. In the real world, laboratories often face difficulties due to lack of samples positive for parasites, such as protozoa. In a recent multicenter Korean study, the overall prevalence of protozoa causing diarrhea was only to 0.8% [12]. Accordingly, accurate pre-use evaluations are difficult to conduct with clinical samples in such a low endemic area. For this reason, there is a lack of data regarding the performance evaluation of the BD MAX EPP in Korea, with only 2 reports available [13,14]. Moreover, in those studies, the number of positive cases (2 E. histolytica, 1 C. parvum) was too small to adequately verify clinical performance. Instead of clinical samples, spiking samples with known parasitic pathogens could serve as alternative materials for assay performance evaluation [15,16]. Defined stool samples spiked with a known amount of target are expected to provide relatively precise and controlled performance. Therefore, this study aimed to verify the performance of the BD MAX EPP using simulated samples in a low endemic setting.
Materials and Methods
Ethics statement
Stool samples for this study were collected in accordance with the guidelines and approval of the institutional review board of Asan Medical Center (No. 2024-0661). Given the nature of the project, a waiver of consent was granted for the remaining samples.
Sample collection
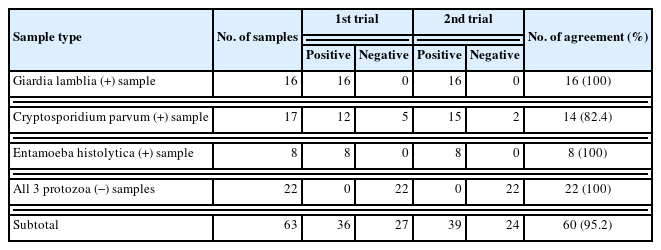
We collected residual stool specimens from patients who visited Asan Medical Center for health check-ups. For the evaluation of E. histolytica, 8 stool samples obtained from a patient with a liver abscess caused by E. histolytica were included. Additionally, 22 stool specimens requested for occult blood testing without definitive evidence of protozoa infection by stool microscopy were used as negative controls, and 17 stool samples from patients with enteritis caused by bacterial or viral pathogens were used for cross-reactivity assessment. We obtained standard materials for 3 protozoa, namely, C. parvum oocysts (P102C, 1×106 oocysts in 4 ml), G. lamblia cysts (P101, 1×106 cysts in 4 ml) from Waterborne Inc. (New Orleans, LA, USA). E. histolytica genomic DNA (5.3×103 copies/100 ml, ATCC 30459D) from ATCC was also obtained for the limit of detection (LoD) assessment. Then, we prepared simulated stool samples to verify the diagnostic performance of the kits. Positive simulated samples for C. parvum and G. lamblia were prepared by spiking the above standard materials to stool samples without definitive evidence of protozoa infection by stool microscopy. Specifically, 6,250 (n=8) and 62,500 (n=9) oocysts of C. parvum and 6,250 (n=7) and 62,500 (n=9) cysts of G. lamblia were used per 1 ml of stool sample. As previously mentioned, to check for cross-reactivity, 17 samples from patients with enteritis caused by bacterial or viral pathogens were tested using the BD MAX Enteric Bacterial Panel (BD Diagnostics, Baltimore, MD, USA) or the BD MAX Enteric Viral Panel (BD Diagnostics, Baltimore, MD, USA), and they showed positive results for the following pathogens: Salmonella spp. (n=2), Campylobacter spp. (n=1), Shigella spp. (n=1), Yersinia spp. (n=1), Plesiomonas spp. (n=1), Vibrio spp. (n=1), enterotoxigenic Escherichia coli (ETEC) (n=1), enterohemorrhagic E. coli (EHEC) (n=1), norovirus (n=3), sapovirus (n=2), rotavirus (n=1), adenovirus (n=1), and astrovirus (n=1), respectively. LoD was determined by diluting each of the 3 standard materials into 6 to 14 concentrations, depending on the organism, and testing each concentration twice. Repeatability was assessed by retesting all positive and negative samples twice. Accuracy was evaluated by comparing the results of the BD MAX system with the intended results in both ways: the first trial data only and the correction data by retesting. Diagnostic performances, such as sensitivity and specificity, were calculated using MedCalc version 8.0 (Medcalc Software, Mariakerke, Belgium).
Results
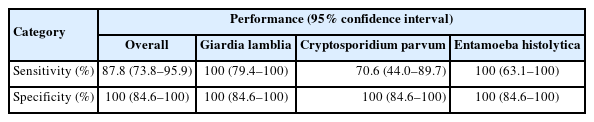
The LoD of the BD MAX EPP assay was determined by serial dilution of standard materials as 781 cysts/ml for G. lamblia, 6,250 oocysts/ml for C. parvum, and 125 DNA copies/ml for E. histolytica (Table 1). The simulated G. lamblia-positive stool samples with concentrations exceeding 6,250 cysts/ml consistently yielded positive results (100% of concordance rate) (Table 2). By contrast, the simulated C. parvum-positive stool samples with the concentration of 6,250 oocysts/ml showed relatively low concordance rates (50% at first trial and 75% after retesting). In the samples with the concentration of 62,500 oocysts/ml, the concordance rates were 89% at first trial and 100% after retesting. Overall agreement was found to be 95.2% (60/63), exhibiting a fair repeatability (Table 3). In the case of C. parvum, a relatively lower agreement of 82.4% (14/17) was observed, which might be caused by the relative insensitivity of this assay. No cross-reactivity to other enteric bacterial or viral pathogens nor interference with blood in the stool samples was observed. Overall diagnostic performances of the assay were determined using the first testing results (Table 4). The sensitivity of the assay (95% confidence intervals) was 87.8% (73.8%–95.9%), specificity 100% (84.6%–100%). For C. parvum, the sensitivity of the assay was 70.6% (44.0%–89.7%), specificity 100% (84.6%–100%).
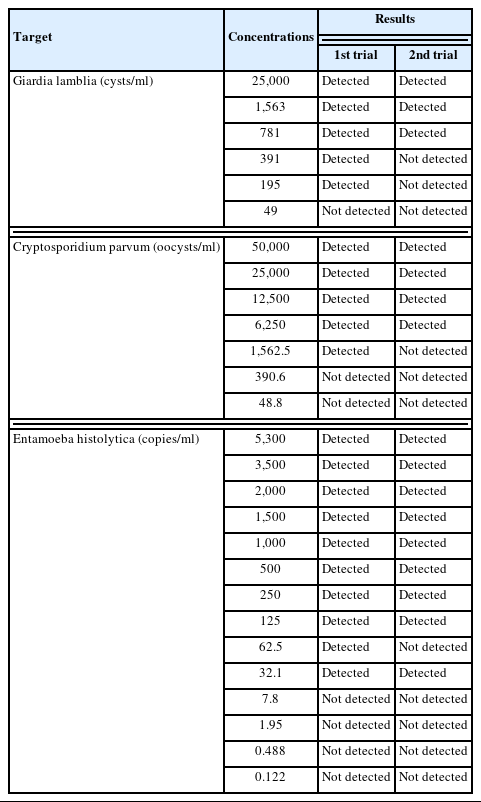
Verification of limit of detection of the BD MAX Enteric Parasite Panel assay using standard materials
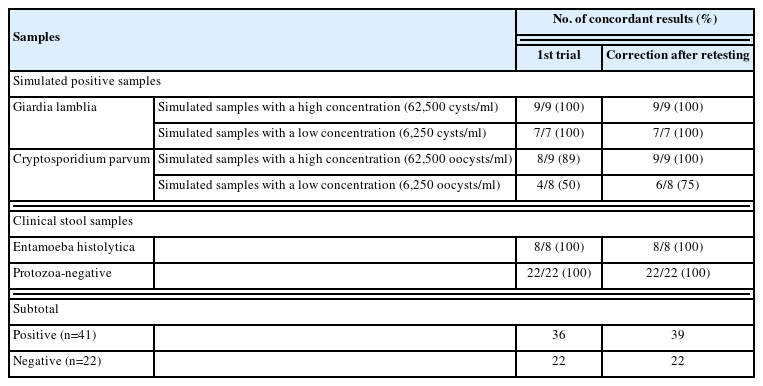
Concordant results of the BD MAX Enteric Parasite Panel assay using simulated and clinical stool samples
Discussion
Previous studies in Korea provided important initial evaluations of the BD MAX EPP using clinical samples [13,14]. In those studies, however, the number of positive samples was too small and even all of 3 targets were not included in their test sets (1 C. parvum [13], 2 E. histolytica [14]), which posed challenges in achieving a fully comprehensive assessment of diagnostic performance. In this study, we expanded upon these findings by using spiked samples to conduct a more controlled evaluation tailored to the low endemic context of Korea. This approach enables a systematic assessment under low-prevalence conditions, addressing gaps that previous research could not fully explore. The LoD measured in our study was higher than the LoD values proposed by the manufacturer for both C. parvum and G. lamblia, which were 10.67 organisms for G. lamblia and 160.17 organisms for C. parvum, as well as 16.79 organisms for E. histolytica. This discrepancy can be attributed to several factors, including differences in specific experimental conditions and the use of simulated samples, which differ from the manufacturer’s ideal testing conditions. Direct comparison with the manufacturer’s value for E. histolytica is difficult because our LoD was determined using standardized ATCC DNA. Numerous evaluations of BD MAX EPP have been conducted in countries outside Asia [6–10]. Previous studies reported 99.0%–100% sensitivities and 96.6%–100% specificities for G. lamblia and 100% sensitivity and specificity for E. histolytica, which are consistent with the results of our study. Another study comparing the EPP with an in-house PCR assay using clinical samples reported a moderate sensitivity of 66.7% for G. lamblia, but this study demonstrated a high sensitivity of 100% for G. lamblia, highlighting a significant difference [6]. This difference may be ascribed to several factors, such as the performance of the kit itself and the study cohort in the studies. It is noteworthy that 70.6% sensitivity and 100% specificity for Cryptosporidium species were obtained in our study, which are lower than the 95.0%–100% sensitivities and 98.9%–100% specificities reported in previous studies. Several factors might have influenced the relatively low sensitivity of C. parvum detection. These include insufficient disruption of oocysts, which might have resulted in the low yield of DNA extraction during simplified extraction method of this assay [17,18]; the use of unpreserved stool samples, which might have affected DNA stability and extraction efficiency; and the possibility that spiked samples with C. parvum do not directly reflect the biological conditions of naturally infected samples. Collectively, the results of this study verified the high sensitivity, specificity, and accuracy of the BD MAX EPP assay for G. lamblia and E. histolytica. Although the assay showed a somewhat lower sensitivity of 70.6% for C. parvum, this level is considered sufficient for clinical use. Thus, this assay could be employed as a first-line tool for routine diagnosis of protozoan infection. In the past, protozoan infection was microscopically diagnosed using special stain or direct fluorescent-antibody stain on preserved concentrated stool specimens [8]. However, this technique is labor intensive and requires well-trained and highly skilled technicians for optimal interpretation. Furthermore, protozoan parasites are difficult to identify especially when they are present in low numbers; thus, microscopic examination is unsuitable for routine diagnosis in regions with a low incidence of protozoan infections [3–5,13]. Even though molecular diagnostics demonstrate better sensitivity or specificity than microscopy, conventional PCR methods, particularly in-house assays, often lack standardization, leading to inconsistent results and complicating interpretation across different laboratories. In contrast, the BD MAX EPP offers advantages, such as automated procedures and the absence of additional nucleic acid extraction steps. In this study, we did not focus on direct comparisons with microscopy or other commercial PCR panels but rather on the validation of the BD MAX EPP as a user-friendly diagnostic tool using expanded simulated samples.
This study had some limitations. Above all, we utilized DNA standard materials instead of E. histolytica live cells, unlike the cell-based evaluations for G. lamblia and C. parvum. Nonetheless, we found that DNA standards can also be considered as a meaningful alternative for the evaluation of diagnostics when regulatory issues restrict access to live cell materials. In Korea, molecular diagnosis for diarrhea-causing protozoa has recently been available within the healthcare reimbursement system. Molecular diagnostics may be further used as a routine diagnostic tool for diarrhea-causing protozoa. In the future, a new era of first-line routine diagnostic process for diarrhea-causing protozoa should be prepared. Although we could not obtain a sufficient number of clinical samples for verification, the BD MAX EPP demonstrated good performance for clinical use. Our findings support the use of spiked samples as a practical alternative for evaluating diagnostic accuracy, helping to establish a reliable primary diagnostic process for protozoan infections in low-incidence settings and contributing to improved public health outcomes.
Notes
Author contributions
Conceptualization: Won EJ
Methodology: Won EJ
Investigation: Won EJ, Park B
Visualization: Won EJ, Park B
Funding acquisition: Won EJ
Project administration: Won EJ
Supervision: Sung H, Kim MN
Writing – original draft: Won EJ, Park B
Writing – review & editing: Won EJ, Park B, Sung H, Kim MN
Conflict of interest
The authors declare no conflict of interest related to this study.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (grant No. 2022 R1C1C1002741).