Detection of trichomonads in induced sputum from asthma patients in Korea
Article information
Abstract
Trichomonads are flagellated protozoa that have occasionally been detected in the human respiratory tract, although detection rates have often been underestimated. We applied a nested PCR assay targeting the 18S rRNA gene of trichomonads to induced sputum from asthma patients to determine the prevalence of Trichomonas. Induced sputum was collected from 41 adults with asthma and analyzed through nested PCR using broad-range trichomonad primers and DNA sequencing for species identification. Nested PCR detected trichomonad DNA in 10 of the 41 (24.4%) samples. Sequencing and phylogenetic analysis revealed Trichomonas tenax in 8 cases and Tetratrichomonas sp. in 2 cases. These findings indicate that trichomonads can be present in the lower airways of patients with asthma, warranting further investigation into their clinical relevance.
Trichomonads are flagellated protozoa primarily found in the human digestive and reproductive tracts but have also been occasionally detected in the respiratory system, particularly in patients with aspiration risk or chronic pulmonary conditions [1–5]. In fact, studies have identified several Trichomonas and related species in human pulmonary samples, including Trichomonas vaginalis, Trichomonas tenax, Pentatrichomonas hominis, and Tetratrichomonas spp. [1,6]. Among these species, T. tenax, a commensal of the human oral cavity, has been most often detected in patients with respiratory infections [5]. Reports of pulmonary or pleural trichomoniasis associated with T. tenax have dated back decades, often in individuals with poor oral hygiene or aspiration risk [7]. In contrast, Tetratrichomonas spp. have been more commonly detected in animals. However, several Tetratrichomonas cases have recently been identified in human respiratory tracts, particularly in individuals with chronic lung diseases [2]. To the best of our knowledge, no previous study has specifically investigated the presence of trichomonads in the respiratory tracts of asthma patients.
We applied the nested PCR protocol targeting the 18S rRNA gene described by Lin et al. [1] to detect trichomonads in induced sputum from 41 adult asthma patients. Genomic DNA was extracted from 200 μl of induced sputum sediment using the FastDNA SPIN kit for soil (MP Biomedicals, Carlsbad, CA, USA). The 18S rRNA gene was first amplified using the primers TRC1-F (5′-GGTAATTCCAGCTCTGCG-3′) and TRC1-R (5′-TGGTAAGTTTCCCCGTGT-3′) under the following conditions: initial denaturation at 98°C for 2 min; 20 cycles of 98°C for 10 sec, 53°C for 30 sec, and 68°C for 30 sec; and a final extension at 68°C for 5 min. Subsequently, a nested PCR was performed using the primers TRC2-F (5′-GTTAAAACGCCCGTAGTC-3′) and TRC2-R (5′-CCAGAGCCCAAGAA CTAT-3′) under the following conditions: initial denaturation at 98°C for 2 min; 35 cycles of 98°C for 10 sec, 54°C for 30 sec, and 68°C for 30 sec; and a final extension at 68°C for 5 min. All patients were outpatients recruited from Severance Hospital, Seoul, Korea between July 2023 and December 2024. Among the included patients, 22, 15, and 4 had severe, moderate, and mild asthma, respectively, based on the treatment steps defined by the Global Initiative for Asthma guidelines. This study was approved by the Institutional Review Boards and Ethics Committees, and the subjects provided written informed consent (IRB No. 4-2023-0296). A phylogenetic tree was constructed using the Neighbor-Joining method with 1,000 bootstrap replicates and computed using MEGA11 software (The Pennsylvania State University, Pennsylvania, PA, USA). Reference sequences were obtained from GenBank (National Center for Biotechnology Information). The sequence data generated from this study have been submitted to GenBank under accession numbers PV533683-PV533692.
Among the 41 patients who underwent PCR testing and sequencing analysis, 10 (24.4%) tested positive. Among the samples that tested positive, 8 (19.5%) were found to have T. tenax, with all sequences showing complete identity, whereas the remaining 2 (4.9%) were found to have Tetratrichomonas spp. Notably, both patients who tested positive for Tetratrichomonas had severe asthma, with not Tetratrichomonas having been detected in those with moderate or mild asthma; however, this association was not statistically significant (Fisher exact test, P=0.49). Phylogenetic analysis confirmed the species-level identity of the detected trichomonads (Fig. 1). In particular, T. tenax-positive sequences clustered tightly with the reference T. tenax sequences from GenBank, showing 100% similarity. The 2 Tetratrichomonas-positive sequences grouped closely with Tetratrichomonas sp. FJ711081.1, a strain previously isolated from a human empyema case, with 99.73% similarity (1 bp difference out of 370 bp, excluding primers) [8].
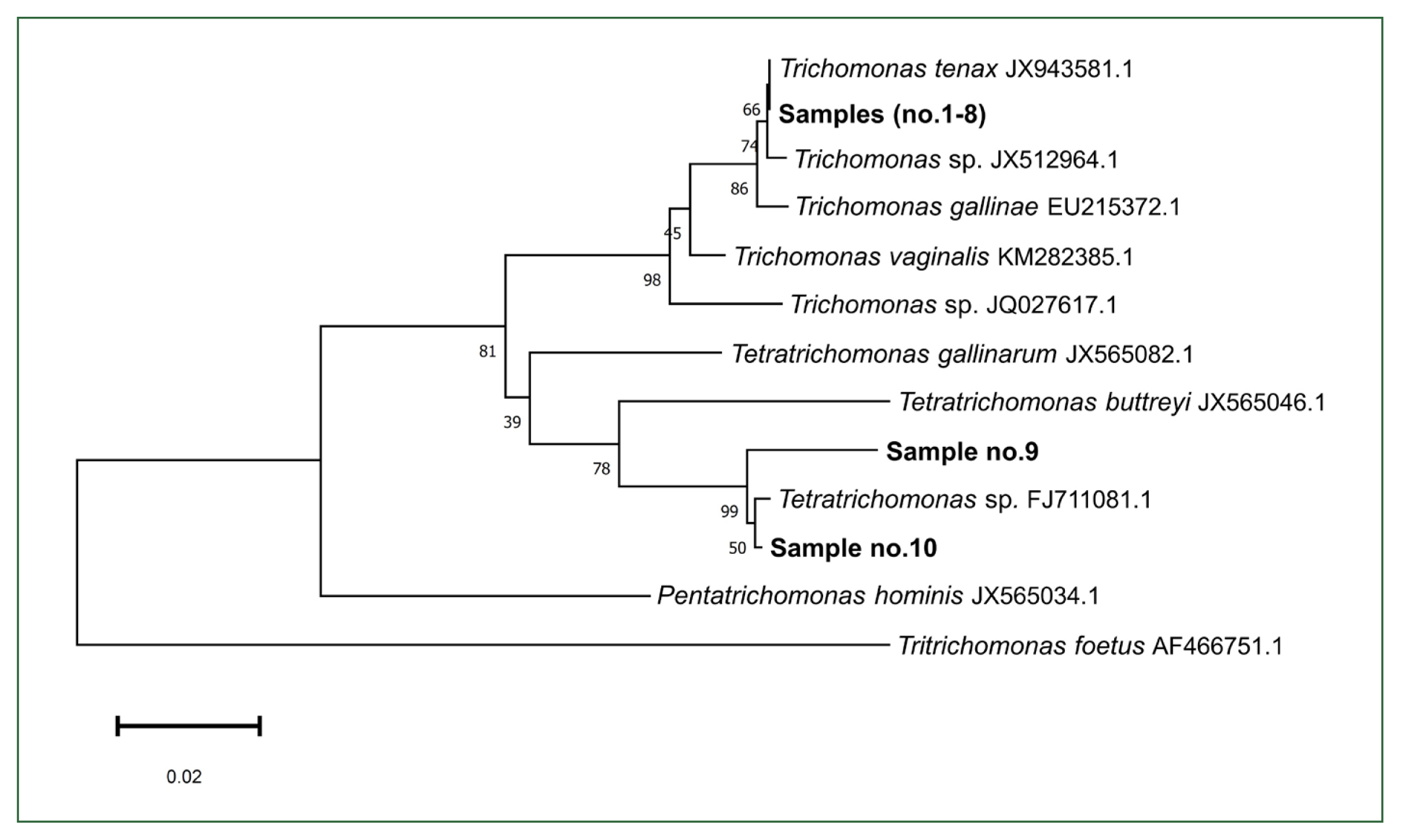
Phylogenetic tree based on 18S rRNA sequences showing the relationship between trichomonads detected in induced sputum samples and reference sequences extracted from GenBank. The tree was constructed using the Neighbor-Joining method with 1,000 bootstrap replicates.
These findings align with the results reported by Lin et al. [1] using bronchoalveolar lavage fluid, which also showed the presence of T. tenax and Tetratrichomonas. The detection of Tetratrichomonas in our patients is noteworthy and raises several possible interpretations. One possibility is the underestimation of zoonotic colonization given that Tetratrichomonas spp., such as Tetratrichomonas buttreyi and Tetratrichomonas gallinarum, are parasites primarily associated with animals [9,10]. Another possibility is the existence of a human-adapted parasitic strain considering the repeated identification of Tetratrichomonas in human respiratory samples and the near-identical sequence identity to a strain previously isolated from human empyema and pyopneumothorax [6,8,11]. Additionally, Kutisova et al. [12] identified several Tetratrichomonas strains from the bronchi of patients with chronic pulmonary diseases. These findings suggest that humans may act as not only incidental hosts but also permissive hosts for an emerging respiratory Tetratrichomonas species.
T. tenax has been frequently detected in the oral cavities of individuals with poor dental hygiene, with a reported prevalence ranging from 12% to 32% [13]. However, its prevalence in the lower respiratory tract remains underexplored due to the limited number of studies [14,15]. Although T. tenax likely originates from the oral cavity and enters the lower airways via microaspiration, the possibility of contamination during sputum collection should also be considered, especially given its high prevalence in the mouth. In the current study, induced sputum samples were collected following the standard sputum induction protocol, which includes precollection oral rinsing to minimize oral contamination, although complete elimination could not be ensured. Recent in vitro studies have shown that T. tenax disrupts epithelial barrier function and induces IL-6 production in gingival epithelial cells while causing no cytotoxicity in pulmonary epithelial cells [16].
One limitation of this study is the lack of a healthy control group. Our samples were collected from asthma patients according to standard protocols used in clinical practice, which restricts direct comparison with healthy controls. Hence, future studies including induced sputum samples from healthy individuals are warranted to better understand the significance of trichomonad detection.
In this study, trichomonads in induced sputum samples collected from patients with asthma were detected using nested PCR targeting the 18S rRNA gene. Further studies are necessary to determine whether these trichomonads promote airway inflammation or asthma pathogenesis or are merely incidental colonizers considering that their pathogenic potential remains speculative given the small sample size and absence of functional or immunological data.
Notes
Author contributions
Conceptualization: Park JW
Data curation: Oh HK
Formal analysis: Choi JH
Methodology: Kim M
Project administration: Yi M
Validation: Cho YH
Writing – original draft: Yi M
Writing – review & editing: Kim JY
Conflict of interest
The authors declare no conflict of interest related to this study.
Acknowledgments
This study was supported by the National Research Foundation of Korea (NRF) funded by the Korea government (MSIT) (Grant No. RS-2024-00456300) and by a grant from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (Grant No. RS-2023-KH139971).
