Distribution and genotypes of Enterocytozoon bieneusi in raccoon dogs in Korea
Article information
Abstract
Enterocytozoon is a genus of microsporidian parasites, with Enterocytozoon bieneusi being a well-known species. It infects various mammalian hosts, including humans, and exhibits zoonotic potential. Out of the 97 fecal and intestinal samples collected from wild raccoon dogs in Korea, 12 (12.4%) tested positive for E. bieneusi via PCR, revealing 2 genotypes: genotype D and EbpA. Both genotypes were found to belong to the zoonotic Group 1. Notably, this study is the first to report the EbpA genotype in Korea. Although studies on E. bieneusi in raccoon dogs are relatively limited, the findings suggest potential public health concerns.
Enterocytozoon is a genus of microsporidian parasites, with Enterocytozoon bieneusi being a well-known species that infects a wide range of mammalian hosts, including humans, and exhibits zoonotic potential [1]. The Microsporidia phylum comprises approximately 1,700 identified species, 17 of which are known to infect humans. Among them, E. bieneusi accounts for over 90% of the reported human microsporidial infections [2]. Initially identified as an opportunistic pathogen in individuals with acquired immune deficiency syndrome, E. bieneusi is now known to cause symptomatic and asymptomatic infections in immunocompromised and immunocompetent individuals [3]. The symptoms typically include diarrhea and lethargy [2].
E. bieneusi is transmitted through the ingestion of spores from contaminated food, and water, or through direct contact with infected sources. Once inside the host, the spores use a polar tube to inject their contents into the epithelial cells of the intestine. The parasite multiplies in the cytoplasm, initially forming multinucleated plasmodia, and then developing into sporogonial stages with complex structures. Sporoblasts form and mature into spores, which are released when the host cell ruptures and are excreted in feces, thereby continuing the transmission cycle [3].
E. bieneusi is primarily detected using molecular and microscopic techniques. PCR, which targets the internal transcribed spacer (ITS) region of the rRNA gene, is considered the most sensitive and specific method. This approach enables detection of isolates and their genotyping for epidemiological studies. To date, over 800 genotypes of E. bieneusi have been identified through polymorphism analysis of the ITS region of the rRNA gene [4]. These genotypes have been classified into at least 11 distinct phylogenetic groups based on ITS sequence data [5]. However, recent studies have suggested that these genotypes can be classified into 12–15 groups [5,6]. Group 1 is the largest phylogenetic group and includes numerous genotypes found in humans and animals, such as genotypes D and Type IV, which have been reported in various countries, including Korea [7].
In Korea, E. bieneusi has been detected in several farm animals, including cattle (genotypes D, Type IV, CEbA, CEbD, CEbF, I, and J), pigs (genotypes PigEBITS3 to PigEBITS5, CAF1, and H), and horses (genotypes horse1 and horse2) [8–10]. It has also been identified in wild animals, such as bats (genotypes KBAT1 to KBAT4, and BEB8), deer, wild boars (genotypes D, H, EbpC, Korea-WL1 to WL2, Korea-WL5 to WL6, KWB1 to KWB4, and RDK), leopard cats (genotype Korea-WL4) [11–13], and shelter cats (genotype Peru11) [14].
Despite these findings, research on the prevalence of E. bieneusi in raccoon dogs in Korea remains limited. To date, only 1 study has investigated the presence of E. bieneusi in raccoon dogs and other wild animals in Korea [15]. To address this gap in the literature, this study aimed to investigate the prevalence of E. bieneusi in wild raccoon dogs in Korea and assess its potential public health significance.
Between May 2017 and October 2023, a total of 97 fecal and intestinal samples were collected from road-killed and trapped wild raccoon dogs across Korea. This study was supported by the Ministry of Environment, Korea. The collection of feces from carcasses was not related to research ethics. Fresh fecal samples were placed in individual tubes, transported in ice-packed containers maintained at 4°C, and delivered to the laboratory for DNA extraction. Relevant sampling information, such as the season and region of collection, was recorded for each sample. Unclear or uncertain data were marked as “unknown.” The sampling regions were categorized based on administrative boundaries into northern (Gangwon and Gyeonggi Provinces) and central/southern (Chungcheong, Gyeongsang, and Jeolla Provinces) areas. DNA extraction was performed using the QIAamp Fast DNA Stool Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. The concentration and purity of the extracted DNA were measured using the Infinite 200 PRO NanoQuant plate reader (Tecan, Männedorf, Switzerland). For amplification, PCR was performed using the AccuPower HotStart PCR Premix (Bioneer, Daejeon, Korea), followed by a nested PCR to target and amplify the ITS region of E. bieneusi for detection purposes [16]. In the first round of PCR, the primers ITSF1 (5′-GGTCATAGGGATGAAGAG-3′) and ITSR1 (5′-TTCGAGTTCTTTCGCGCTC-3′) were used as the forward and reverse primers, respectively.
In the second round, ITSF2 (5′-GCTCTGAATATCTATGGCT-3′) served as the forward primer and ITSR2 (5′-ATCGCCGACGGATCCAAGTG-3′) as the reverse primer [17]. PCR amplification was performed using the Mastercycler Pro (Eppendorf, Hamburg, Germany). The thermal cycling conditions included an initial denaturation step at 95°C for 5 min, followed by 35 cycles of denaturation at 95°C for 30 sec, annealing at 55°C for 30 sec, and extension at 72°C for 30 sec. A final extension was performed at 75°C for 5 min.
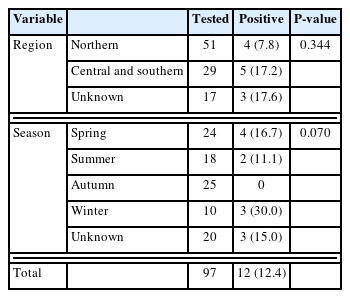
Among the 97 specimens collected, 12 were positive for the E. bieneusi ITS region (Table 1), resulting in a prevalence of 12.4%. This prevalence was lower than that reported in other animals in Korea, such as wild animals (45.2%), calves (16.9%), cattle (14.9%), and pigs (14.2%), but higher than that in bats (2.6%) and Korean cats (Jeju Island, 3.8%) [11]. Studies in China reported a prevalence ranging from 2.6% to 28.1%, whereas a study in Poland reported a prevalence of 40.2% [2]. A previous study conducted in Korea considered raccoon dogs as wild animals and reported a prevalence of 35.4% [15]. These studies demonstrate that the prevalence can vary depending on factors such as the geographical region, sample size, and collection period (season or date). Previous studies on raccoon dogs in China have reported variable prevalence rates. In Shandong Province (eastern China), no statistical significance observed based on region, gender, or age, whereas in northern China, significant associations were observed with region and farm scale, but not with the collection year, age, or presence of diarrhea [2,18].
In this study, prevalence of E. bieneusi was analyzed by region and season. The northern region had a prevalence of 7.8%, whereas the central and southern regions had a prevalence of 17.2%. By season, the prevalence of E. bieneusi was 30.0% in winter, 16.7% in spring, and 11.1% in summer, with no positive cases observed in autumn (Table 1). Statistical analysis using the chi-square test was conducted with the IBM SPSS Statistics version 26 (IBM Corp., Armonk, NY, USA). This analysis showed no statistically significant associations between E. bieneusi prevalence and with season or regions, and the association between temperature and region was not clearly established. A clear association between the prevalence of E. bieneusi and a specific region could not be confirmed due to the limited number of samples tested for each region and season. This aspect needs to be assessed in follow-up studies.
Four of the 12 PCR-positive samples (2 from feces and 2 from intestinal tissue) were sequenced at Macrogen (Daejeon, Korea). Two aligned sequences were generated using BioEdit version 7.2.5 and submitted to GenBank under the accession numbers PV483842 to PV483845. Phylogenetic analysis, including sequence comparison, was conducted using the NCBI Web BLAST tool (http://www.ncbi.nlm.nih.gov/blast).
A phylogenetic tree was constructed using MEGA7, with bootstrap analysis based on 1,000 replicates. All sequences were confirmed to cluster within Group 1, which is known to be zoonotic (Fig. 1). Among the 4 sequences obtained, 3 (RDF-19-ITS, RDS-28-ITS, and RDS-29-ITS) were identified as genotype D (PV483843 to PV483845), whereas 1 (RDF-17-ITS) was classified as genotype EbpA (PV483842). For genotype comparison and tree construction, the reference sequences included a genotype D sequence from raccoon dogs in China (KU847361) and a genotype EbpA sequence from a dog in China (KJ668725). The genotype D (PV483843 to PV483845) sequence identified in this study showed 99.96% identity with a sequence identified in a previous study on raccoon dogs (LC436511) and 100% identity with sequences from a Chinese raccoon dog (KU847361), a Korean water deer (LC436451), and a human (AF101200). The genotype EbpA (PV483842) sequence identified in the present study exhibited 99.7% identity with a sequence from a Chinese dog (KJ668725) and 100% identity with a sequence from a Swiss pig (AF076040).
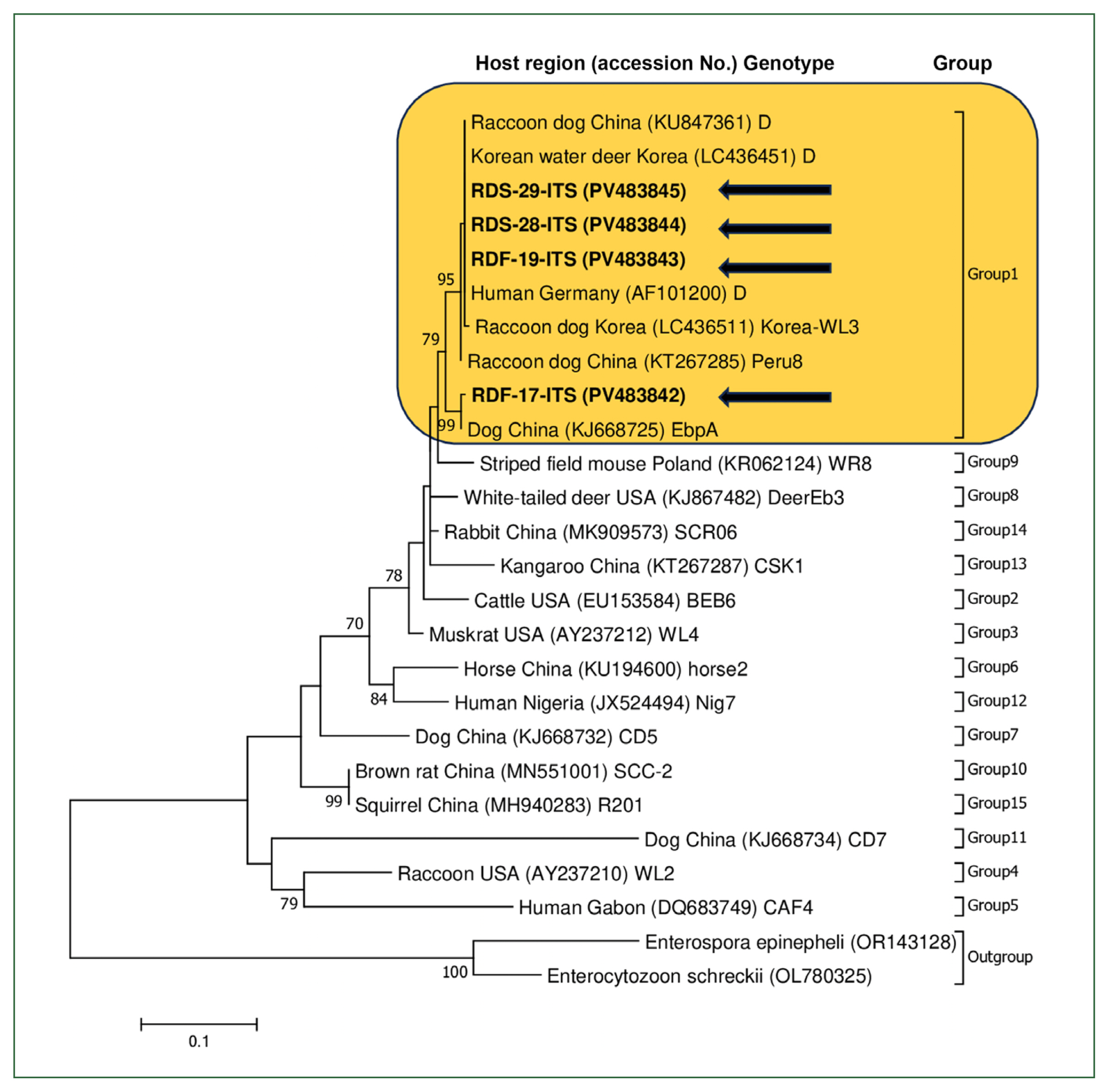
A phylogenetic analysis of Enterocytozoon bieneusi identified in raccoon dogs from Korea was conducted based on the internal transcribed spacer (ITS) region. The phylogenetic tree was generated using the maximum likelihood method in MEGA7, with bootstrap analysis performed using 1,000 replicates. Arrows indicate the sequences obtained. Each sequence includes information on the host species, country of origin, and the corresponding GenBank accession number. Genotypes and their respective groups and subgroups are labeled on the right side of the tree.
Group 1 is the most diverse genotypic group characterized by low host specificity and contains several subgroups (1a–1i) [7]. It is zoonotic and has been identified in various animals, including raccoon dogs. Approximately 20 different E. bieneusi genotypes have been identified in raccoon dogs, most of which belong to the zoonotic Group 1, such as D, Type IV, Peru8, and EbpC [4,18].
Genotype D was first identified in raccoon dogs in 2 studies conducted in China, suggesting that raccoon dogs could serve as a potential reservoir for the zoonotic E. bieneusi genotype [19,20]. This genotype was also identified in raccoon dogs in Korea [15], including in the current study. The genotype EbpA was first detected in raccoon dogs in a study from China [16]. This genotype has also been identified in humans, with previous studies reporting its presence in various regions and populations [7,20]. However, to date, there has been no evidence of the EbpA genotype in Korea. This study is the first to confirm its presence in the country. These findings underscore the zoonotic potential of the EbpA genotype and its relevance to human health.
Raccoon dogs are among the most commonly found wild animals in Korea observed near urban parks. In this study, the E. bieneusi sequences identified in raccoon dogs showed 100% identity with sequences previously reported in humans from other countries, suggesting that raccoon dogs may serve as a potential zoonotic source of infection. This study provides a nationwide analysis of E. bieneusi in fecal and intestinal samples from wild raccoon dogs in Korea. Its significance lies in identifying the EbpA genotype, which had not previously been reported in Korea. Like genotype D, EbpA is classified within Group 1 and is considered to have zoonotic potential. Therefore, ongoing research is essential to collect additional samples and expand the geographic scope of surveillance, thereby supporting efforts to monitor the risk of zoonotic transmission through human contact.
Notes
Author contributions
Conceptualization: Kwak D
Data curation: Park HM, Lee H, Kwak D
Formal analysis: Park HM, Lee H
Investigation: Park HM, Lee H, Chae SJ, Son K, Lee S
Methodology: Park HM, Lee H
Project administration: Kwak D
Supervision: Kwak D
Validation: Park HM, Lee H, Chae SJ, Son K, Lee S, Nazim K, Lee SH, Koo Y, Kang J, Seo MG, Park SJ, Rhee MH
Visualization: Park HM, Lee H
Writing – original draft: Park HM, Lee H
Writing – review and editing: Chae SJ, Son K, Lee S, Nazim K, Lee SH, Koo Y, Kang J, Seo MG, Park SJ, Rhee MH, Kwak D
Conflict of interest
Dongmi Kwak serves as an editor of Parasites, Hosts and Diseases but had no involvement in the decision to publish this article. No other potential conflicts of interest relevant to this study were reported.
Acknowledgments
This study was supported by the National Institute of Wildlife Disease Control and Prevention under a research support project (1800-1833-345).