Identification and confirmation of SUMOylation-modified proteins in Giardia trophozoites
Article information
Abstract
Posttranslational modification by the small ubiquitin-related modifier (SUMO) is one of the crucial cellular processes in Giardia lamblia, a protozoan pathogen. In this study, 5 candidate SUMO substrate proteins of G. lamblia trophozoites were chosen based on their enrichment through affinity chromatography using a SUMO-interaction motif: never in mitosis A-related kinase (NEK), aminoacyl-histidine dipeptidase (AHD), protein disulfide isomerase 2 (PDI2), alcohol dehydrogenase 3, and ornithine carbamoyltransferase. Transgenic Giardia trophozoites expressing 1 of the 5 candidate SUMO substrate proteins were constructed, and their expression was confirmed by western blot using hemagglutinin-tag. Arginine deiminase (ADI) protein was expressed in Giardia trophozoites as a positive control. Cell extracts were processed for affinity chromatography using SUMO-interaction motif resin. As expected, the SUMOylated form of ADI was detected in the affinity chromatography extracts of ADI-expressing cells. Among the 5 candidate proteins, SUMOylated forms of NEK, AHD, and PDI2 were identified in the affinity chromatography extracts. These results suggest that NEK, AHD, and PDI2 activity is modulated via SUMOylation in Giardia trophozoites.
The posttranslational modification (PTM) of proteins is a dynamic process that efficiently modulates cellular response to various stimuli in eukaryotes. Ubiquitination is one of the main PTMs involved in controlling proteasome-mediated degradation of various proteins, such as cell cycle-related components, and coordinating the localization and activity of diverse proteins. As an alternative mode of ubiquitination, the covalent and reversible attachment of small ubiquitin-like modifier (SUMO) to lysine residues of target proteins regulates cellular functions such as nuclear-cytoplasmic transfer, DNA repair, cell cycle regulation, and transcriptional regulation [1].
Giardia lamblia, a pathogen causing diarrheal disease, is present as 2 distinct forms, trophzoites and cysts. Trophozoite is a multiplying form found in the host’s small intestine, whereas cyst is an infective form found outside the host. Interestingly, Giardia lacks the anaphase-promoting complex required for the ubiquitination-dependent degradation of cell cycle components [2]. Therefore, SUMOylation is considered an important PTM involved in regulating Giardia trophozoites division.
The G. lamblia genome contains genes encoding SUMO, SUMO-activating enzyme, SUMO-conjugating enzyme, and SUMO nuclease, 2 genes have been predicted as encoding SUMO ligases [3]. Two known substrates of Giardia SUMO are α-tubulin [4] and arginine deiminase (ADI), a metabolic enzyme that converts arginine to citrulline [5].
We aimed to define the role of SUMOylation in Giardia by identifying SUMOylation-modified proteins. G. lamblia trophozoites (ATCC 30957; American Type Culture Collection, Manassas, VA, USA) were cultured at 37°C in modified TYI-S-33 medium supplemented with 10% heat-inactivated calf serum (Gibco, Rockville, MD, USA), and 0.75 mg/L bovine bile (Sigma-Aldrich, St. Louis, MO, USA). Approximately 4×107 Giardia trophozoites were harvested by centrifugation at 1,900×g for 20 min at 4°C. After washing twice with 1×phosphate-buffered saline. One hundred microliters lysis buffer (150 mM NaCl, 200 mM iodoacetamide, 50 mM Tris-HCl (pH 7.5)) with a 1×protease inhibitor cocktail (GenDEPOT, Katy, TX, USA) was added, and the cells were broken down using a sonicator (Qsonica, Newtown, CT, USA).
The resultant extracts were processed via an affinity chromatography using a SUMO-QAPTURE-T kit (Enzo, Farmingdale, NY, USA). Briefly, 100 μg of protein was mixed overnight at 4°C with 40 μl of SUMO-interaction motif (SIM)-conjugated matrix suspension. After 2 washes with binding buffer (50 mM Tris-HCl [pH 7.5]), the bound proteins were eluted with 5×SDS-PAGE loading buffer (250 mM Tris-Cl (pH 6.8), 15% SDS, 50% glycerol, 25% β-mercaptoethanol, 0.01% bromophenol blue). The eluted proteins, along with the extracts used for chromatography and the flow-through fraction, were separated on a 12% SDS-PAGE and then visualized with Quick Coomassie Stain (Protein Ark, Rotherham, UK) (Fig. 1A). These proteins were also reacted with anti-human SUMO-1 antibodies (Fig. 1B). Among the immunoreactive protein bands in the eluted fraction (Fig. 1B), 5 upregulated immunoreactive proteins were excised from the corresponding gel stained with Coomassie blue (Fig. 1A) for subsequent identification at PROTIA (Seoul, Korea). The trypsin-digested products of the 5 chosen proteins were processed via liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) and underwent UniProt (https://www.uniprot.org/) analyses based on the G. lamblia database (GiardiaDB, https://giardiadb.org/giardiadb/app). For each sample, proteins with a Mascot ion score greater than 25 and comparable molecular weight were selected to determine whether they were modified by SUMO (Table 1). The analysis showed the proteins in decreasing size, serially numbered and presented with the GiardiaDB ID members, putative protein functions, predicted molecular weights, and isoelectric points, and information related to LC-MS/MS analysis (Mascot score, matches, sequences, and coverages). The proteins were identified as never in mitosis A-related kinase (NEK) 15409, aminoacyl-histidine dipeptidase (AHD), protein disulfide isomerase (PDI) 2, alcohol dehydrogenase (ADH) 3, and ornithine carbamoyltransferase (OCT).
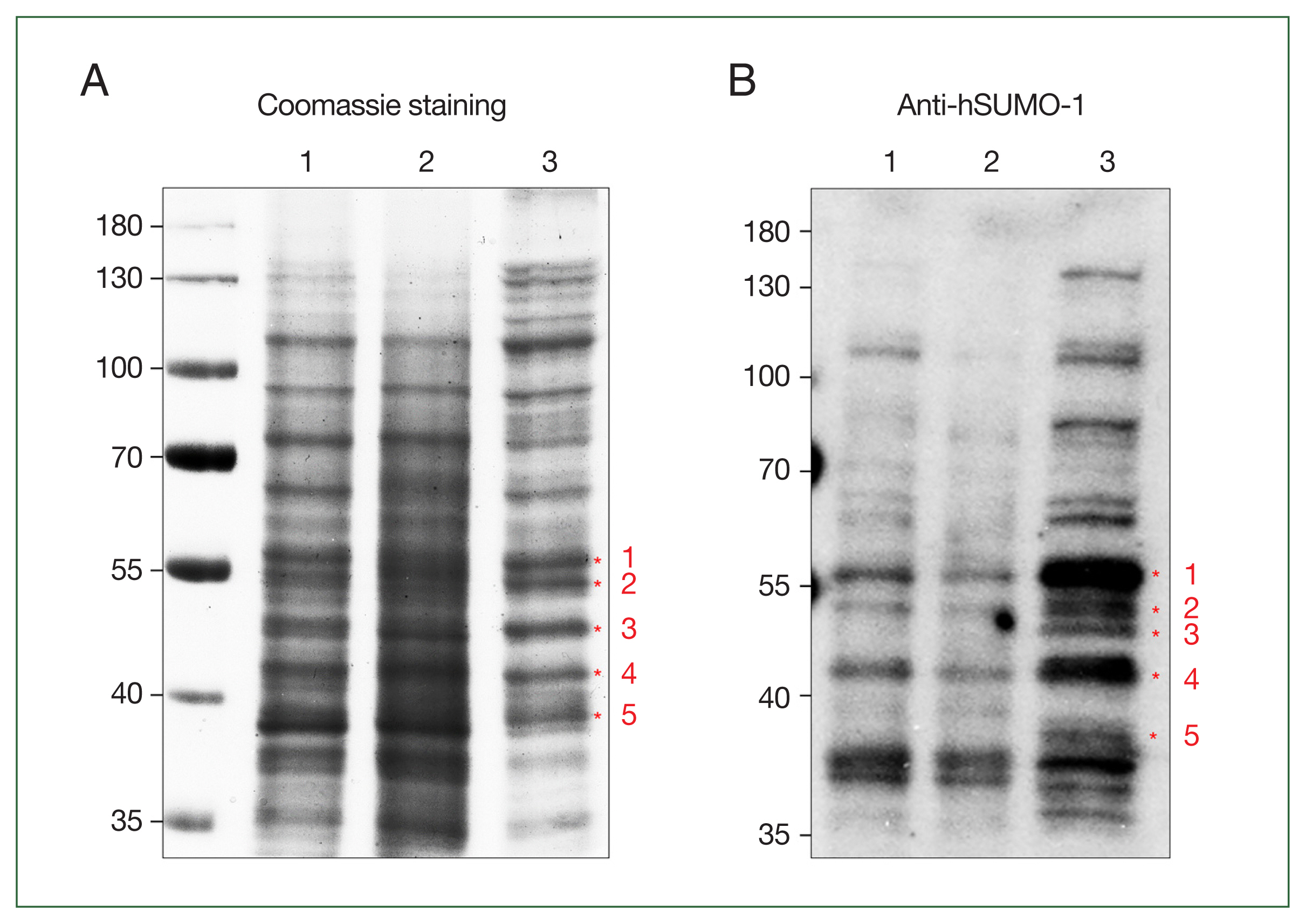
Identification of small ubiquitin-like modifier (SUMO)-modified target proteins using affinity chromatography with SUMO-interaction motif (SIM)-conjugated resin. Giardia extract (100 μg) was reacted with a suspension of the SUMO-interaction motif–conjugated matrix. The extracts used for chromatography (lane 1), the flow-through fraction (lane 2), and the eluted protein (lane 3) were loaded on SDS-PAGE, and the proteins were visualized following Coomassie staining (A). The same proteins were reacted with anti-human SUMO (hSUMO)-1 antibodies (B). The numbered proteins were analyzed by trypsin digestion and subsequent liquid chromatography coupled with tandem mass spectrometry. The identified proteins are listed in Table 1.
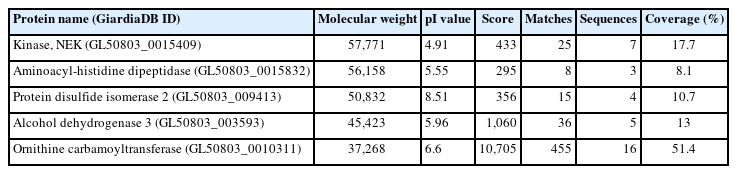
Proteins Identified as candidate small ubiquitin-related modifier substrates using liquid chromatography coupled with tandem mass spectrometry
To determine whether the identified proteins underwent SUMOylation in vivo, i.e., within Giardia trophozoites, transgenic Giardia were constructed in which 1 of the 5 proteins was expressed as a hemagglutinin (HA)-tagged form following transfection with an expression plasmid. Trophozoites expressing HA-tagged ADI were included as controls. Table 2 presents a list of primers and plasmids used in this study [6]. To construct the plasmid expressing ADI, an ADI DNA fragment with its own promoter (GL50803_112103) was created using PCR with Giardia genomic DNA (gDNA) and primers t112103-F-infusion and t112103-R-infusion. The PCR amplicon was subsequently cloned into pKS-3HA.NEO digested with NotI using an In-Fusion HD Cloning Kit (Takara, Shiga, Japan), generating pADI-HA.NEO. A 1,545-bp DNA fragment encoding the NEK15409 open reading frame (GL50803_15409) and its 200-bp promoter was amplified from Giardia gDNA by PCR using primer pair t15409-F/r15409-R. A DNA fragment containing the promoter and open reading frame of PDI2 (GL50803_9413) was also prepared by PCR amplification using the primer pair t9413-F/r9413-R. The resultant NEK15409 and PDI2 DNA fragments were cloned into the HindIII and XhoI sites of pKS-3HA.NE O to produce p15409-HA.NEO and p9413-HA.NEO, respectively. DNA fragments encoding AHD (GL50803_15832) and ADH3 (GL50803_3593) were generated by PCR using the primer pair t15832-F/t15832-R and t3593-F/t3593-R, respectively. The resultant DNA fragment was inserted into the SacII and NotI sites of pKS-3HA.NEO to generate p15832-HA.NEO and p3593-HA.NEO, respectively. A DNA fragment encoding OCT (GL50803_10311) was obtained from Giardia gDNA by PCR using the primers t10311-F-infusion and t10311-R-infusion and then ligated with pKS-3HA.NEO using an In-Fusion HD Cloning Kit.
These plasmids were transfected into Giardia trophozoites as previously described [6]. Transfected Giardia cells were selected in TYI-S-33 medium supplemented with 600 μg/mL G418. The expression of HA-tagged proteins was confirmed by western blot using anti-HA antibodies. The expression of HA-tagged ADI was detected in extracts of Giardia carrying the expression plasmid for ADI. One of the 2 immunoreactive bands was 57 kDa, the expected size of HA-tagged ADI. Additionally, a larger band (~72 kDa) was consistently observed (Fig. 2A), and it was thought that this may be SUMOylated ADI, as detected in a previous study [5]. The Giardia trophozoites harboring the empty vector showed no immunoreactive bands with anti-HA antibodies. In contrast, the amounts of G. lamblia protein disulfide isomerase 1 (PDI1, GL50803_29498) were equivalent in cells with the vector or ADI-expression plasmid. Giardia harboring the expression plasmid for NEK15409, AHD, PDI2, ADH3, and OCT were examined for the expression of the HA-tagged protein via western blotting using anti-HA antibodies (Fig. 2B, C, D, E, and F, respectively). The expression of these proteins was absent in the cells carrying the empty vector. The amount of PDI1 was monitored as a loading control for relative protein expression in the transgenic G. lamblia.
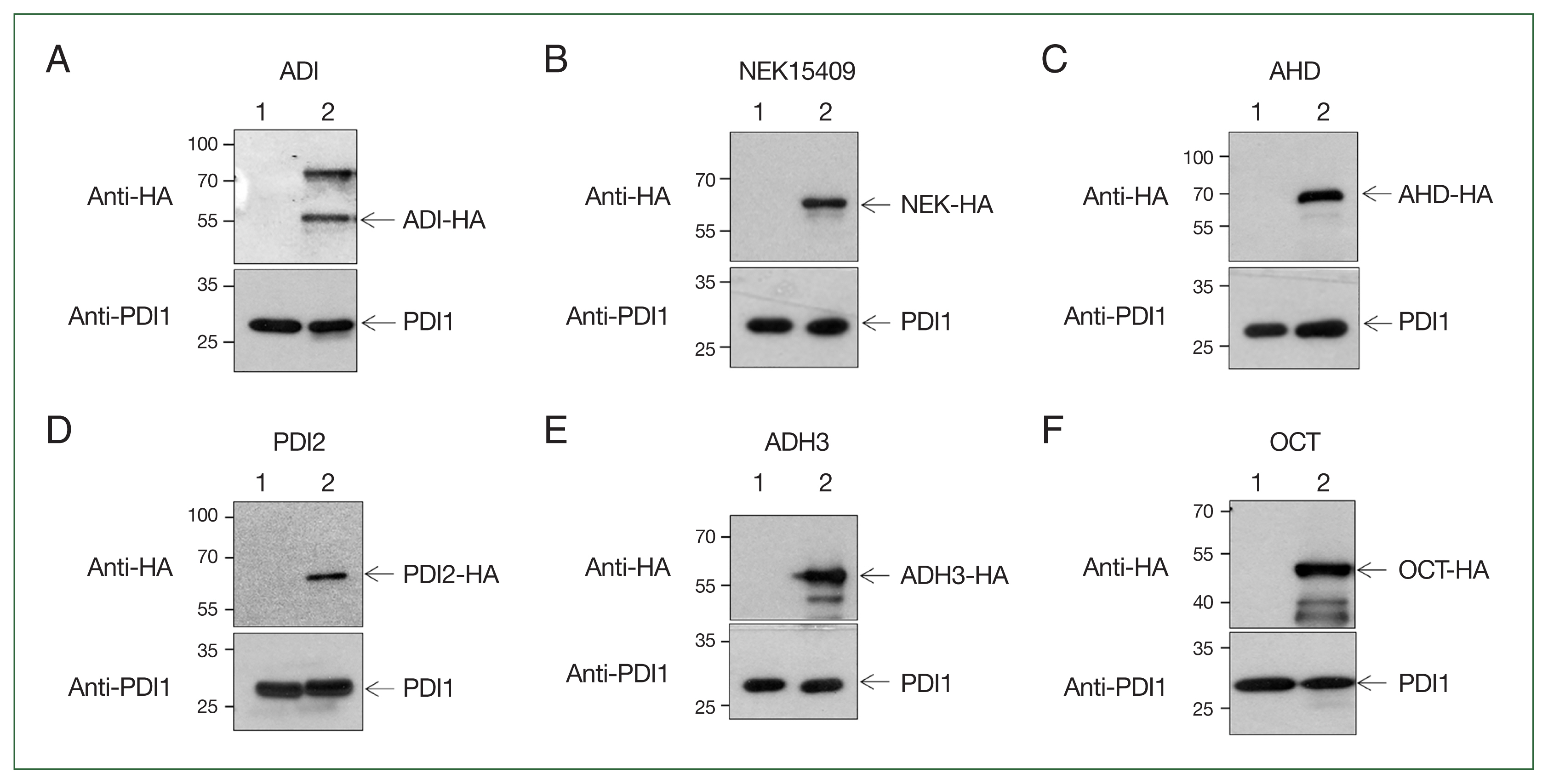
Expression of hemagglutinin (HA)-tagged small ubiquitin-like modifier target proteins. Expression plasmids for the 6 selected proteins were transfected into Giardia. The transfectants were selected by culture G418-containing medium. Extracts from Giardia carrying the vector plasmid (lane 1) or expression plasmids (lane 2) were reacted with anti-HA antibody. After stripping, the same blot was incubated with anti-protein disulfide isomerase 1 (PDI1) (GL50803_29487) as a loading control. Giardia expressing arginine deiminase (ADI) (A), NEK15409 (B), aminoacyl-histidine dipeptidase (AHD) (C), protein disulfide-isomerase 2 (PDI2) (D), alcohol dehydrogenase 3 (ADH3) (E), and ornithine carbamoyltransferase (OCT) (F).
Giardia expressing 1 of the 6 HA-tagged proteins (ADI, NEK15409, AHD, PDI2, ADH3, and OCT) were used as input samples for affinity chromatography conjugated with SIM. Flow through and bound proteins were monitored as unbound and eluent samples, respectively, for further studies. Western blot analysis using anti-HA antibodies revealed that the fractions of HA-tagged ADI-expressing Giardia contained both modified and unmodified ADI (Fig. 3A). The SUMOylated form of the protein was larger than the unmodified protein. Interestingly, an immunoreactive band with a higher molecular weight than the unmodified protein was detected in Giardia expressing HA-tagged NEK15409, AHD, and PDI2 (Fig. 3B, C, and D, respectively). In contrast, no immunoreactive bands were observed in the experiments involving ADH3 and OCT (Fig. 3E and F, respectively).
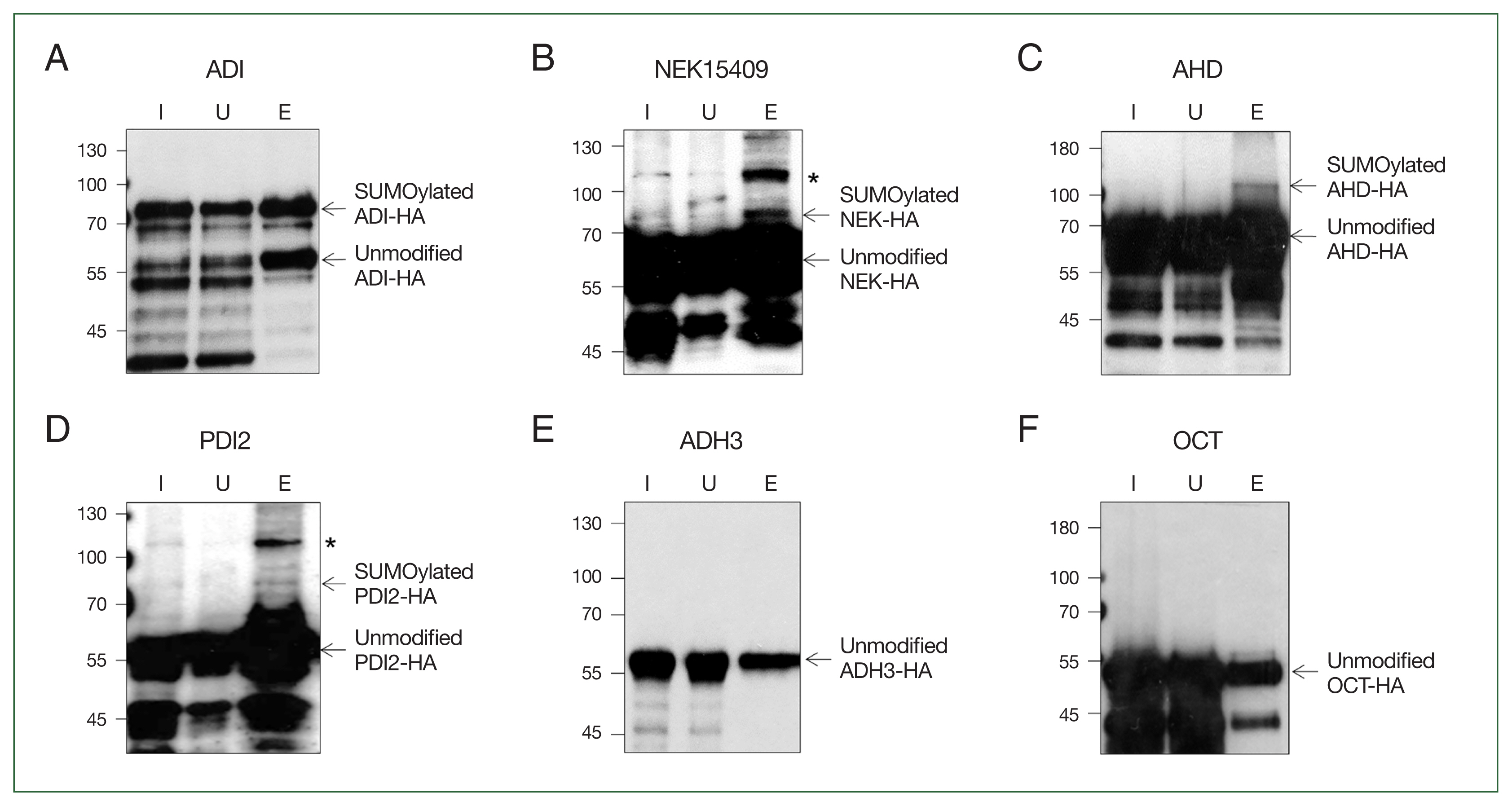
Confirmation of proteins modified by SUMOylation via an affinity chromatography of transgenic Giardia expressing candidate proteins. Giardia extract expressing hemagglutinin (HA)-tagged protein was reacted with a SUMO-interaction motif–conjugated matrix suspension. Fractions for input extract (lane I), unbound (lane U), and eluent (lane E) were loaded onto SDS-PAGE and reacted with anti-HA antibody. Arginine deiminase (ADI) (A), NEK15409 (B), aminoacyl-histidine dipeptidase (AHD) (C), protein disulfide-isomerase 2 (PDI2) (D), alcohol dehydrogenase 3 (ADH3) (E), and ornithine carbamoyltransferase (OCT) (F). Asterisks indicate nonspecific bands in the elution fraction.
SUMOylation is one of the PTMs present in G. lamblia. Because this organism lacks anaphase-promoting complex [2], it is more likely that SUMOylated proteins play a main role in diverse cellular processes, including cell cycle control. RNA interference-mediated depletion of SUMO causes defects in the shape of Giardia trophozoites [4]. Two centrins of G. lamblia have been identified as SUMO substrates, and defects in SUMOylation resulted in the arrest of cell cycle progression and conformational changes in Giardia (Yeo et al., unpublished result). In Aspergillus, a mutation in UbaB, a SUMO-activating enzyme, resulted in abnormal chromatin bridges indicating the role of SUMO in chromosome segregation [7]. In budding yeast, SUMOylation of septin is critical for recruiting Fir1, a cytokinesis checkpoint protein, at the hourglass stage of septin prior to its transformation into the double-ring form [8]. Mutagenesis of nuclear and spindle-associated protein 1 in humans revealed that 2 independent SUMOylation events affected its nuclear localization and activity by causing differential association with RANBP2, a nuclear porin [9].
Among the known SUMO substrates in Giardia, ADI was included as a control in the SUMOylation assay (Fig. 3A). As expected, formation of the larger immunoreactive HA-tagged ADI (modified ADI) was the most obvious compared with other candidate SUMO substrates, namely NEK15409, AHD, and PDI2 (Fig. 3B, C, and D, respectively). ADI is a multifunctional protein. As a metabolic enzyme, it is involved in the arginine dihydrogenase pathway with carbamate kinase and OCT. This pathway is important not only for energy generation but also for immune evasion of this protozoan against host cells [5]. Interestingly, we identified OCT as a SUMO substrate when performing affinity chromatography using SIM (Fig. 1A; Table 1). However, SUMOylation of OCT was not verified through subsequent detection from Giardia expressing HA-tagged OCT (Fig. 3F). Therefore, it is unlikely that SUMOylation modulates the activity of this pathway. Besides its metabolic role, ADI plays a multifaceted role via its peptidyl-ADI activity, and it modifies the conserved CRGKA tail of variant-specific surface proteins [5].
One of the SUMO substrates identified in this study was NEK15409 (Fig. 3B). NEK is a conserved cell cycle-related kinase in eukaryotes. Interestingly, the Giardia genome has almost 200 NEKs, but information on their expression, activity, and function is lacking in most cases. Two NEKs, NEK1/NEK92498 (GL50803_92498) and NEK2/NEK5375 (GL50803_ 5375), have been investigated as the main NEKs, demonstrating roles in morphogenesis, cytokinesis, and excystation of G. lamblia [10]. Four NEKs (NEK1, NEK2, NEK16279, and NEK101534) were identified as components of basal bodies, adhesive disc or flagella of Giardia trophozoites [11]. RNA-seq analysis of G1/S-arrested versus G2/M-arrested Giardia revealed that 3 NEKs (NEK1, NEK5999, and NEK95593) were upregulated at the G2/M phase [12]. Furthermore, several transcriptomic analysis have shown the upregulation of 8 NEKs (NEK8350, NEK11364, NEK91451, NEK95593, NEK3957, NEK114307, NEK15409, and NEK133701) during encystation [13]. In addition, defects in NEK8445 altered microtubule polymerization, eventually leading to structural malformation of flagella and adhesive discs in Giardia trophozoites [14]. NEK15409, one of the encystation-induced NEKs was identified as a SUMO substrate in this study, suggesting that PTM modulates its activity and possibly localization.
PDI is a representative marker protein of the endoplasmic reticulum. Its thioredoxin-like domain contains a cysteine redox-active site, facilitating the formation of disulfide bonds in proteins, resulting in the refolding of secretory or denatured proteins [15]. Five PDIs have been identified in Giardia, all which have a single thioredoxin-like domain in the N-terminus region: PDI1 (GL50803_29487, 26 kDa), PDI2 (GL50803_9413, 50 kDa), PDI3 (GL50803_14670, 13 kDa), PDI4 (GL50803_103713, 39 kDa), and PDI5 (GL50803_8064, 15 kDa) [16]. In Giardia, PDI2 appears to play a major role in disulfide bond reduction, as only PDI2 was able to complement the activity of the knockout mutant Escherichia coli [16]. Observations of PDI1, 2, and 3 in the endoplasmic reticulum and encystation specific vesicles suggest their role in the assembly and release of cysteine-rich variable surface proteins in trophozoites and cyst wall formation during encystation [17]. Interestingly, PDI4 and PDI2 bind to nitazoxanide, suggesting their potential as drug targets [18].
The last candidate SUMO substrate is AHD (GL50803_9413) (Fig. 3C). AHD is a metallopeptidases performing various functions including protein degradation, protein maturation, and cell cycle control. In Spironucleus vortens, a diplomonad fish parasite closely related to Giardia, AHD was found to bind to nitroimidazole, causing redox imbalance due to antioxidant failure [19]. This enzyme is secreted from Giardia trophozoites upon exposure to human intestinal epithelial cells [20]. Our finding of AHD as a SUMO substrate (Fig. 3C) suggests that secretion of AHD is regulated by SUMOylation.
The identification of NEK15409, AHD, and PDI2 as candidate SUMO substrates indicates that their activity and localization are modulated via SUMOylation in Giardia trophozoites. The roles of these components in division or/and pathogenesis will be upcoming issues in study on G. lamblia.
Notes
Author contributions
Conceptualization: Kim J
Formal analysis: Kim J
Funding acquisition: Park SJ
Investigation: Yeo HR, Shin MY
Methodology: Yeo HR, Shin MY
Supervision: Kim J, Park SJ
Visualization: Kim J, Park SJ
Writing – original draft: Kim J, Park SJ
Writing – review & editing: Kim J, Park SJ
Conflict of interest
The authors declare no conflict of interest related to this study.
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Korean government (MSIT) (NRF-2023R1A2C1002475).