Genotype analysis of Cryptosporidium spp. prevalent in a rural village in Hwasun-gun, Republic of Korea
Article information
Abstract
Two species of Cryptosporidium are known to infect man; C. hominis which shows anthroponotic transmission between humans, and C. parvum which shows zoonotic transmission between animals or between animals and man. In this study, we focused on identifying genotypes of Cryptosporidium prevalent among inhabitants and domestic animals (cattle and goats), to elucidate transmittal routes in a known endemic area in Hwasun-gun, Jeollanam-do, Republic of Korea. The existence of Cryptosporidium oocysts was confirmed using a modified Ziehl-Neelsen stain. Human infections were found in 7 (25.9%) of 27 people examined. Cattle cryptosporidiosis cases constituted 7 (41.2%) of 17 examined, and goat cases 3 (42.9%) of 7 examined. Species characterizations were performed on the small subunit of the rRNA gene using both PCR-RFLP and sequence analysis. Most of the human isolates were mixtures of C. hominis and C. parvum genotypes and similar PCR-RFLP patterns were observed in cattle and goat isolates. However, sequence analyses identified only C. hominis in all isolates examined. The natural infection of cattle and goats with C. hominis is a new and unique finding in the present study. It is suggested that human cryptosporidiosis in the studied area is caused by mixtures of C. hominis and C. parvum oocysts originating from both inhabitants and domestic animals.
INTRODUCTION
Cryptosporidium is a small protozoan parasite found in the brush-border of mucosal epithelia of various species of mammals, birds, reptiles, and fishes(Morgan et al., 1999). After Tyzzer (1907) found Cryptosporidium muris in the gastric gland of mice, more than 20 Cryptosporidium species have been proposed based on their host specificities, oocyst morphologies, and genetic characteristics (Morgan et al., 1999; Xiao et al., 2004). Human infection cases have been reported in Cryptosporidium parvum (including C. hominis), C. meleagridis, C. muris (renamed as C. andersoni), and C. felis, but most cases were caused by C. parvum (Morgan et al., 2000a; Xiao et al., 2001) or C. hominis (Morgan-Ryan, 2002). Cryptosporidiosis is known to be an opportunistic parasitosis of humans (Widmer, 1998) with a worldwide distribution (Griffiths, 1998). This disease occurs in 5-50& of AIDS patients (Spano and Crisanti, 2000), and an important cause of AIDS-associated deaths due to severe diarrhea (Widmer et al., 1998).
Several studies have reported the characteristics of C. parvum with respect to its transmission, and 2 genotypes of C. parvum were suggested to exist in humans (Peng et al., 1997; McLauchlin et al., 2000). One was named as the human genotype, i.e., Cryptosporidium hominis, which is transmissible between humans, and the other was a bovine genotype, redesignated as C. parvum, which is transmissible between man and vertebrates, especially cattle (Morgan-Ryan et al., 2002). Several studies had revealed the differential characteristics between C. hominis and C. parvum particularly in terms of genetic differences, whereas others have focused on its pathogenic and epidemiologic properties (Okhuysen et al., 1999; Morgan et al., 2000a; Eisenberg et al., 2005). The different transmission cycles of C. parvum and C. hominis between man and animals could allow us to trace sources of oocyst contaminations.
In the Republic of Korea, the first documentation of the presence of Cryptosporidium was done by a case report of chicken cryptosporidiosis (Mo et al., 1988) and subsequently C. parvum was reported in mice (Chai et al., 1990). Thereafter, human C. parvum infections were reported in epidemiological studies (Chai et al., 1996, 2001; Seo et al., 2001). In particular, a study in a small rural village of Hwasun-gun verified the existence of C. parvum in both humans and cattle, suggesting that the responsible parasite might be the bovine genotype (Chai et al., 2001). However, genetic approaches of Cryptosporidium in this village have not been progressed.
The present study was thus focused on investigating Cryptosporidium species by analyzing genotypes in the known endemic village of Hwasun-gun, by applying PCR-RFLP and sequence analysis of the small subunit (SSU) rRNA gene of Cryptosporidium oocysts collected from man and domestic animals.
MATERIALS AND METHODS
The area studied was a small rural village; Ssangbong-ri, Iyang-myon, Hwasun-gun, Jeollanam-do, a well-known endemic area of cryptosporidiosis (Chai et al., 2001). The previous Cryptosporidium oocyst positive rate was 57% (Chai et al., 2001). In December 2000, fecal samples were collected from 27 people, 17 domestic cattle, and 7 domestic goats living in the village. The samples were mixed well with 2 volumes of 2.5% K2Cr2O7 and stored at 4℃ to maintain oocyst vitality. The specimens were examined for the presence of oocysts, which was confirmed by modified Ziehl-Neelsen staining. Oocysts were then purified using Arrowood and Sterling (1987)'s discontinuous sucrose and CsCl gradient methods. Collected oocysts were re-suspended in 1 ml of 2.5% K2Cr2O7 and stored at -70℃ until required. Genomic DNA was extracted as previously described (Kim et al., 1992).
For species-specific identifications, primer sets were designed based on the SSU rRNA gene locus. Primer sequences unique to Cryptosporidium species, as identified by Xiao et al. (2001), were used in a nested PCR protocol. The primer sets used for the primary PCR were 5'-TTCTAGAGCTAATACATGCG-3' and 5'-CCCTAATCCTTCGAAACAGGA-3' and those used for the secondary PCR were 5'-TTCTAGAGCTAATACATGCG-3' and 5'-CCCATTTCCTTCGAAACAGGA-3'. For restriction fragment analyses, 5 µl of the second PCR products were digested for 4 hr in 20 µl reaction mixtures containing 10 U of Ssp I (Roche, Mannheim, Germany) or 10 U of Vsp I (Gibco BRL, Grand island, New York, U.S.A.), as recommended by the manufacturers. Digested mixtures were loaded on 2% agarose gels and visualized by ethidium bromide staining after electrophoresis. Amplified PCR products in agarose gels were extracted using the QIAGEN Gel Elution Kit (QIAGEN K.K., Tokyo, Japan).
PCR products eluted from gels were directly sequenced using a model ABI Prime 377 Automatic Sequencer (Perkin Elmer, Foster City, California, U.S.A.), when they were suitable for sequence analyses. However, if eluted products were too small or unsuitable for direct sequencing, the pGEM®-T Easy Vector Systems I (Promega, Madison, Wisconsin, U.S.A.) was used. All sequences were aligned and analyzed using Megalign (DNA Star, Madison, Wisconsin, U.S.A.) and Sequence Navigator (Applied BioSystems, California, U.S.A.) programs.
RESULTS
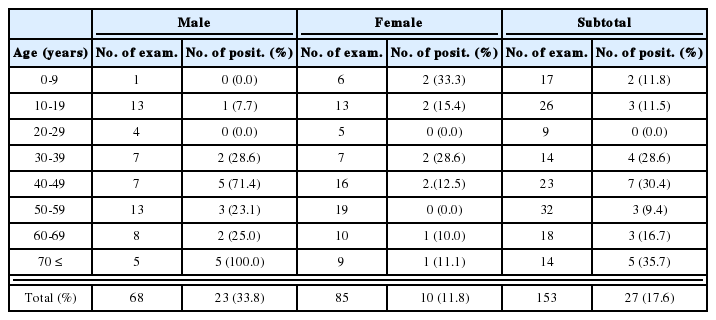
Cryptosporidium oocysts were found in 7 human fecal specimens (25.9% in positive rate), 7 cattle (41.2%), and 3 goats (42.9%) (Table 1), and these were encoded serially (Fig. 1). In most cases of smeared samples, the number of oocysts did not exceed 20 per smear, and oocyst sizes ranged 4-6 µm for both C. parvum and C. hominis. No other protozoan cysts were detected, but unidentified helminth eggs were observed in 2 goats. A sucrose gradient separation method and a CsCl gradient method were found to be useful for purifying oocysts from fecal samples; however, oocyst losses were incurred at each step.

Prevalence of Cryptosporidium spp. among people and domestic animals in the study area, Ssangbong-ri, Iyang-myon, Hwasun-gun, Jeollanam-do

Gel electrophoresis of nested PCR-RFLP based on the small subunit (SSU) rRNA gene of Cryptosporidium. Stained with ethidium bromide on 2% agarose gel. The species of Cryptosporidium was determined by digestion with endonuclease, Ssp I (B) or Vsp I (C), of nested PCR products (A). Most of isolates had both C. hominis and C. parvum patterns (B, C). The white arrowhead (ca. 628 bp) is C. parvum-specific and the black arrowhead (ca. 556 bp) indicates C. hominis-specific bands obtained by Vsp I (C) digestion. M, DNA size marker; lanes 1-5, isolates from villagers; lanes 6-11, isolates from domestic cattle; lanes 12-14, isolates from domestic goats.
Nested PCR using genomic DNA, extracted from purified oocysts, was sufficient for endonuclease digestion (Fig. 1A). However, the amplification failed for 1 isolate from a man (data not shown) even though a small number of oocysts were detected by modified Ziehl-Neelsen staining. Digestion with Ssp I fragmented the second PCR products (ca. 830 bp) into 3 bands (448, 247, and 106 bps) (Fig. 1B), which is characteristic for both C. parvum and C. hominis (Xiao et al., 1999), except in 2 samples, one each from a man and a cattle (data not shown). Digestion of the PCR products with Vsp I produced 3 bands, 2 of which (ca. 628 and 104 bps) are specific for C. parvum, and 2 (ca. 556 and 104 bps) for C. hominis (Xiao et al., 1999) (Fig. 1C).
The sequences of the PCR products were analyzed in 5 human isolates (sample Nos. 1, 2, 3, 4, and 5; Fig. 1), 6 cattle isolates (6, 7, 8, 9, 10, and 11), and 3 goat isolates (12, 13, and 14). The PCR products from sample 1 were sequenced directly and others were sequenced using the cloned PCR products. A few variations were observed at a repeated thymine locus. However, sequences in the polymorphic regions (positions 260-267, 637-653, and 680-699) were all C. hominis type (Table 2), and no sequences for C. parvum type were obtained. Sequences of C. hominis from humans, cattle, and goats were accessed in GenBank under accession numbers DQ054817 (human isolate), DQ054818 (cattle isolate), and DQ054819 (goat isolate).
DISCUSSION
As shown by the results, oocysts of both C. hominis and C. parvum genotypes were identified in humans by PCR-RFLP analyses. Isolates from domestic animals also showed the same species genotypes, although sequencing of the PCR products successfully revealed only C. hominis genotype. Obtaining only C. hominis sequence may suggest predominance of C. hominis among the Cryptosporidium oocysts in the study area. However, the co-infection of both C. hominis and C. parvum in man and animals is a unique finding in the present study.
Human cryptosporidiosis, whatever the parasite species, is caused by direct contact with contaminated food or through drinking water. Ingestion of only small numbers of viable Cryptosporidium oocysts could cause human cryptosporidiosis (Smith et al., 1993). In the study area, water is supplied from a storage reservoir used by both villagers and domestic animals. Swimming and fishing are prohibited in the reservoir; however, a few domestic animals including cattle and goats are allowed to roam freely around the reservoir. Thus, if the reservoir was contaminated with Cryptosporidium oocysts from these animals, villagers and their animals could be easily exposed to oocysts. Sylvatic transmissions of C. parvum involving deer and smaller mammals have also been suggested to be important sources of human cryptosporidiosis (Perz and Le Blancq, 2001).
In the waterborne outbreaks in England, human cryptosporidiosis was found to be primarily caused by drinking water contamination and C. parvum was identified as the causative parasite species (McLauchlin et al., 2000). The area studied in the present study showed a prevalence and seasonality (Chai et al., 2001) similar to those reported in England (McLauchlin et al., 2000). However, in the present study, it seems that C. hominis genotype predominated, as shown by the sequencing data. In this respect, water reservoir contamination by infected animals may not have been the main cause of human cryptosporidiosis. We suggest that cryptosporidiosis in the studied village may have been caused by food handlers directly contacting foods contaminated by oocysts, i.e., during the preparation of family meals and animal foods.
A unique finding in the present study was the presence of C. hominis genotype in domestic cattle and goats. Of 7 cattle isolates, 6 contained C. hominis genotype as shown by PCR-RFLP (Fig. 1C), and all of the 3 goat isolates contained the same species of Cryptosporidium. Observations in other countries have identified species of Cryptosporidium in bovines and ovines to be C. parvum (McLauchlin et al., 2000). This was an important reason why C. parvum and C. hominis were proposed to be distinct species in terms of transmittal and genetic characteristics (McLauchlin et al., 2000; Morgan-Ryan et al., 2002). However, several recent studies have demonstrated the possibility of animal infections with C. hominis; the examples of animals include interferon-γ knock-out (GKO) mice, immunosuppressed ICR mice, gnotobiotic piglets, a captive primate, a lamb, and calves (Widmer et al., 2000; Giles et al., 2001; Akiyoshi et al., 2002; Guk et al., 2004). Dugong dugon, a marine mammal, was also found infected with the human genotype (Morgan et al., 2000b). In the present study, most of the villagers and their domestic animals, including cattle and goats, reside at the same premises, and thus share a limited biological environment, which may have been responsible for their common infections with C. hominis.
Another possibility is that the Korean isolate of Cryptosporidium might be a third genotype, not discriminated by SSU rRNA gene analyses, i.e., isolates which have genetic characteristics of C. hominis type but have an ability to infect domestic animals. In this respect, successful passages of a human isolate of Cryprosporidium, a mixture of the human and bovine genotypes, to calves and then ICR mice were reported in the Republic of Korea (Guk et al., 2004). If these 2 genetically similar Cryptosporidium species were admixed for a sufficiently long period, genetic recombination could occur. In fact, an experimental meiotic recombination occurred between human (= C. hominis) and bovine genotypes (= C. parvum) in the sexual phase, and produced a progeny that infected GKO mice (Feng et al., 2002).
The results of the present study demonstrated that all of the human, cattle, and goat isolates of Cryptosporidium studied were mixtures of C. parvum and C. hominis. Further studies are required to determine further characteristics of the 2 Cryptosporidium species existing in the Republic of Korea.
Notes
This study was supported by a grant (04-2003-038-01) from the Seoul National University College of Medicine Research Fund (2003) and BK21 Human Life Sciences, Ministry of Education, Republic of Korea.