Biological and Genetic Characterization of Cryptosporidium spp. and Giardia duodenalis Isolates from Five Hydrographical Basins in Northern Portugal
Article information
Abstract
To understand the situation of water contamination with Cryptosporidium spp. and Giardia spp. in the northern region of Portugal, we have established a long-term program aimed at pinpointing the sources of surface water and environmental contamination, working with the water-supply industry. Here, we describe the results obtained with raw water samples collected in rivers of the 5 hydrographical basins. A total of 283 samples were analyzed using the Method 1623 EPA, USA. Genetic characterization was performed by PCR and sequencing of genes 18S rRNA of Cryptosporidium spp. and β-giardin of Giardia spp. Infectious stages of the protozoa were detected in 72.8% (206 of 283) of the water samples, with 15.2% (43 of 283) positive for Giardia duodenalis cysts, 9.5% (27 of 283) positive for Cryptosporidium spp. oocysts, and 48.1% (136 of 283) samples positive for both parasites. The most common zoonotic species found were G. duodenalis assemblages A-I, A-II, B, and E genotypes, and Cryptosporidium parvum, Cryptosporidium andersoni, Cryptosporidium hominis, and Cryptosporidium muris. These results suggest that cryptosporidiosis and giardiasis are important public health issues in northern Portugal. To the authors' knowledge, this is the first report evaluating the concentration of environmental stages of Cryptosporidium and Giardia in raw water samples in the northern region of Portugal.
INTRODUCTION
Infections by the apicomplexan protozoa Cryptosporidium spp. and the flagellate Giardia duodenalis are widespread in humans and animals [1]. The complex life cycle of the former includes cycles of asexual and sexual reproduction in the enterocytes of the intestinal mucosa of the definitive host; mammals and birds [2]. The life cycle of Giardia spp. in the lumen of the duodenal region of the intestine is simpler and includes 2 main stages; trophozoites and cysts [3]. The infectious stages, i.e., oocysts and cysts, are released in the environment through the hosts' feces. Ingestion of both stages from the environment by drinking water or eating raw vegetables is the main transmission route to humans and animals [1,4-6]. Among the 19 species of Cryptosporidium in vertebrates other than fish, Cryptosporidium parvum and Cryptosporidium hominis are most commonly associated with human diseases [5,7]. The species C. parvum is recognized as a zoonotic species. The species Giardia duodenalis is comprised of several genotypes or assemblages; assemblage A and B genotypes are infectious for humans, whereas assemblages C, D, and E are infectious for animal species, including livestock, in a host-specific manner [4,8,9]. The above mentioned species of the genus Cryptosporidium and the genotypes of G. duodenalis have been described as important pathogens in contaminated drinking water, due to 2 main reasons: 1) their resistance and biological viability under the conventional drinking water treatment conditions; 2) the occurrence of cryptosporidiosis and giardiasis outbreaks associated with the consumption of contaminated water [10-12].
To the authors' best knowledge, no cryptosporidiosis or giardiasis outbreaks associated with contaminated water consumption have been described in Portugal. However, clinical cases among immunocompetent patients are well known [13-17]. Both clinical situations are probably underestimated in the country due to the lack of systematic diagnosis of these parasites.
Significant concentrations of the infectious stages of both parasites have been found in water samples collected from rivers in the southern region of Portugal [15,18]. Also, studies on human and animal biological reservoirs indicated presence of important zoonotic species [13,14,16,19]. Systematic evaluation of cattle, surface waters, and humans for the presence of Cryptosporidium spp. and G. duodenalis as well as its genetic characterization using molecular tools are fundamental steps to better understand the epidemiology of the infection and to allow the implementation of risk analysis models for those infections. The present study concerns with the status of Cryptosporidium spp. and G. duodenalis infectious stages in raw water samples of the rivers of northern regions of Portugal.
MATERIALS AND METHODS
The northern region of Portugal
In the north of Portugal, there are 5 major hydrographical basins forming the most important water resources of the country (Fig. 1). These hydrographical basins are named after the main rivers; (1) Minho, (2) Lima, (3) Cávado, (4) Ave, and (5) Douro (Fig. 2). Cávado and Ave rivers run entirely inside national (Portuguese) borders, while Minho, Lima, and Douro are international rivers, with sources in Spain.
Geographic location of the North of Portugal and its 5 hydrographical basins into Iberian Peninsula (same figure was published in Almeida et al. Korean J Parasitol 2010; 48: 43-48).
Water samples
Raw water samples were collected twice a year between January 2004 to December 2006 from 97 sources, including main rivers and respective affluents, from upriver to downriver. The volume of each sample ranged from 25 to 100 L. Samples were filtered through Filta-Max filters (IDEXX Laboratories, Inc., Westbrook, Maine, USA) with a pump on the inlet side of the filter according to the recommendation of the manufacturer. Intact filters were kept in refrigerated containers and transported immediately to the laboratory. The filter was taken from the container and processed with the aid of a Filta-Max Manual Wash Station (IDEXX Laboratories, Inc.) for further elution and concentration process, which consisted of decompression of the the filter, passing the sample through a membrane, and centrifugation. A sample pellet (around 2 ml) was obtained and transferred to a Leighton tube for subsequent immunomagnetic separation (IMS).
Parasite detection
The IMS procedure was performed according to the USEPA method 1623 [20]. Briefly, anti-Giar and anti-Cryptosporidium magnetic beads were mixed with SL Buffer A and SL Buffer B in each Leighton tube containing the sample concentrate (Dynabeads GC-Combo, Invitrogen Dynal, A. S., Oslo, Norway) and incubated 1 hr at room temperature. Then, using 2 magnetic particle concentrators, beads were collected, washed, and transferred into a 1.5 µl tube. Fifty-µl of 0.1 N HCl were added to each sample to dissociate beads from the target organisms, the beads were rejected, and the suspension was transferred to the wells of the slides containing 5 µl of 1.0 N NaOH. The samples were air-dried overnight and stained with FITC-conjugated anti-Cryptosporidium spp. and anti-Giardia spp. monoclonal antibodies, according the manufacturer's instructions (Crypto/Giardia Cells, Cellabs, Sydney, Australia). The excess of FITC-mAb was removed by adding 100 µl of PBS to each well, leaving the slides for 5 min, and aspirating the excess of PBS. A 50-µl aliquot of 4'-6'-diamino-2-phenylindole (DAPI) solution (0.4 µg/ml in PBS) was introduced into each well. The slides were left at room temperature for 15 min, and excess DAPI solution removed by washing the slides in PBS. Slides were examined by epifluorescence microscope. Giardia cysts and Cryptosporidium oocysts were indentified and counted based on their shape and size using a Nikon Optiphot fluorescence microscope (Nikon Corporation, Tokyo, Japan). The number of cysts and oocysts per each well was recorded and concentrations extrapolated per 10 L of sample. Positive and negative controls were performed as indicated by the manufacturer and recommended in the Method 1623.
The mean recovery percentages of oocysts of Cryptosporidium spp. and cysts of Giardia spp. using the Filta-Max system and IMS procedures from water samples is, according to the manufacturer, 50 ± 13% and 41 ± 79%, respectively [21].
DNA extraction, PCR and sequencing
PCR analysis was performed in the samples with the highest density of infectious stages of both parasites detected by DFA. The criterion utilized was the detection of a minimum of 100 cysts/oocysts of any parasite in the total sample volume. In this context, the genetic characterization was executed in 80 samples. The cover slip was separated from the slide and with the aid of cotton swab soaked with 100 µl of distillated water, the surface of the slide was scraped in order to collect the sample. It was confirmed, under microscope observation that the slide had no remaining cysts or oocysts. The tip of the cotton swab was cut and placed in a 1.5 ml tube for subsequent DNA extraction with a QIAamp DNA Mini Kit (QIAGEN GmbH, Hilden, Germany), according to the manufacturer's instructions.
For determining the species of Cryptosporidium and Giardia present in the samples, PCR analyses were performed. A 2-step nested PCR was performed to amplify a portion of the small-subunit (SSU) ribosomal RNA gene of Cryptosporidium spp. [22]. For the molecular typing of Giardia spp., a semi-nested PCR was performed to amplify a portion of the β-giardin gene [23]. For all PCR reactions, negative and positive controls were prepared, with sterile water and reference DNA, respectively. The PCR products were analyzed in agarose gel (1.4%) stained with ethidium bromide under UV light. Images were captured with a gel documentation system (GelDoc2000, BioRad, Hercules, California, USA). The PCR products of the successful reaction were purified by Wizard SV Gel and PCR Clean-up System (Promega, Madison, Wisconsin, USA) and sequenced in both strands by an external laboratory (EUROFINS MWG OPERON, Ebersberg, Germany). Chromatograms were examined with the software ChromasPro (http://www.technelysium.com.au/ChromasPro.html) and the sequences with the software ProSeq (http://www.biology.ed.ac.uk/research/institutes/evolution/software/filatov/proseq.htm). Sequences were compared with the GenBank database with the tool BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and deposited in the database Zoop-Net of the Med-Vet-Net network (http://www.medvetnet.org/cms/).
RESULTS
IMS and DFA detection of infectious stages of Cryptosporidium spp. and G. duodenalis
The number of validated raw water samples in this study was 283. Environmental stages of the protozoa were detected in 72.8% (206 of 283) of the water samples, including 15.2% (43 of 283) positive for cysts of G. duodenalis, 9.5% (27 of 283) for oocysts of Cryptosporidium spp., and 48.1% (136 of 283) for both parasites. In Fig. 3, the percentages of positive and negative samples from the 5 hydrographical basins are shown individually. The Ave basin showed the highest percentage of positive samples; 90.2% of the samples were positive. Minho basin showed the lowest percentage of positive samples, even though this value was more than 64%. In all the 5 hydrographic basins, the co-presence of Cryptosporidium and Giardia counted for the majority of positive samples, with the exception of Minho basin in which Giardia positive samples were slightly more than Cryptosporidium and Giardia positive samples. In the cases where both parasites were present in the same sample, the number of G. duodenalis cysts always outnumbered Cryptosporidium spp. oocysts. We also found no correlation between the concentrations of both parasites, meaning that, when the concentration of Giardia cysts is high, Cryptosporidium oocysts are not necessarily high (data not shown). Furthermore, the range of the concentrations of G. duodenalis cysts was much higher than Cryptosporidium spp. oocysts (0.17-50,000 cysts per 10 L and 0.2-726.1 oocysts per 10 L, respectively). In positive water samples, no empty (i.e., without internal characteristics, or ghosts) or DAPI negative Cryptosporidium spp. oocysts or G. duodenalis cysts were found.
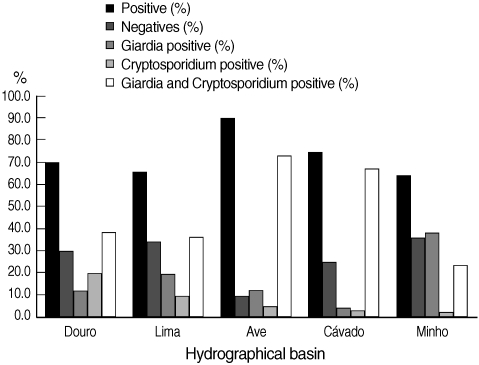
Distribution of the results obtained by Method 1623 EPA, USA, for infectious stages of Cryptosporidium spp. and Giardia duodenalis in water samples collected in 5 hydrographical basins.
In all cases, it was possible to observe an increase of parasite load from upriver to downriver. The majority of water samples from the international rivers (Minho, Lima, and Douro) collected at the border with Spain was negative.
Genetic characterization of species and genotypes isolated
By PCR, it was unable to amplify DNA extracted from slides containing less than 100 oocysts of Cryptosporidium and less than 100 cysts of Giardia. Furthermore, positive amplifications over 3 replicates were never obtained when the number of cysts and oocysts was less than 1,000 per slide. With this criterion, of all the positive IMS samples, PCR amplification was performed over 80 samples.
Genetic characterization was successful in 8 samples for G. duodenalis and 20 samples for Cryptosporidium spp. In 59 samples, PCR amplification was not successful. A summary of the PCR results is shown in Table 1. Cryptosporidium andersoni was found in 16 samples, Cryptosporidium parvum in 2 samples, Cryptosporidium hominis in 1 sample, and Cryptosporidium muris in 1 sample. G. duodenalis assemblage A-II was found in 4 samples, assemblage B in 1 sample, and in the remaining 3 samples assemblages A, B, and E were found.
DISCUSSION
The results of the present study indicate that the infectious stages of Cryptosporidium spp. and G. duodenalisare widely distributed in the rivers of northern Portugal in very significant concentrations. Curiously, G. duodenalis cysts always outnumbered Cryptosporidium spp. oocysts (data not shown). The region has a high density of livestock farms favoring the cycle of parasite amplification. The surface water collected from the rivers is used as drinking water for the animals or used for agricultural purposes, and the feces are directly released into the rivers or reach it by runoff waters.
Systematic studies on the genetic characterization of both protozoans indicate that the genus Cryptosporidium includes species that are infective for humans only (anthroponotic), humans and animals (zoonotic), and other pathogenic species that are not infective for humans [5]. Also, several G. duodenalis genotypes are infective to humans (zoonotic genotypes), and others are non-pathogenic [4]. Amplification and sequencing of genes 18S SSU rRNA for Cryptosporidium spp. and β-giardin for G. duodenalis has been used to identify the zoonotic species and genotypes of the parasites [1,24]. Obviously, it is largely recognized that there is lack of consensus about genetic markers for the correct assignment of the species and subspecies of Cryptosporidiumand Giardia. The gene markers (18S SSU rRNA and β-giardin) are generally accepted as good markers mainly because they are multicopy genes (18S SSU rRNA), restricted to these parasites (β-giardin), and with fixed differences among Cryptosporidium and Giardia species and subspecies (both genes). In our study, in an attempt to produce relevant and comparable results, the choice of 18S SSU rRNA and β-giardin genes, frequently used by the most recognized researchers, was considered.
C. parvum (zoonotic) and C. hominis (anthroponotic) are the most common human-infecting species reported in river water samples in Europe, and C. andersoni is the animal-infecting species [4,25,26]. The same studies indicated G. duodenalis assemblage A as the most common zoonotic genotype, and G. duodenalis assemblage E as the most common non-zoonotic genotype [10,13,17,25,26].
Our results suggest that the contamination of the surface waters in the north of Portugal is highly significant. We have found the zoonotic species of the genus Cryptosporidium described by other authors [18]. In addition, there was higher concentration of G. duodenalis detected by genotyping, with a greater genetic diversity. Assemblage A was found in 7 PCR positive samples (1 A-I and 6 A-II). The presence of the assemblage A-I has been suggested as an indicator of water contamination by livestock, while assemblage A-II has been considered a potential indicator of water contamination by humans [4,13]. Nevertheless, in the northern part of the country assemblage A-I have been found in human samples [17], and assemblage A-II in bovine samples [19,27]. Also, G. duodenalis assemblage B was detected in 4 of 8 samples. Assemblage B has been reported as a zoonotic genotype. The presence of infectious stages of this genotype in water samples has been attributed to water contamination by humans [4]. Assemblage E was detected in 2 samples associated with assemblage A and B, suggesting a mixed human and animal source of contamination.
C. parvum was detected in 2 of the 20 processed samples. This species has a great zoonotic potential, and may have an animal or human source of contamination. A few studies concerning the biological reservoir (human and bovine) in the north of Portugal have indicated C. parvum as an important pathogen infecting the great majority of bovines as well as immunocompromised human patients [14,19]. Recent data suggest that subgenotyping tools may generate more information about the zoonotic potential of C. parvum isolates, although there is still lack of evidences on the usefulness of the generated data for risk assessment [5]. C. hominis, considered an anthroponotic (human-restricted) species, was detected in 1 sample. This species was also reported in water samples and in human stool samples from Portugal [14,18,28]. C. andersoni, a strictly bovine pathogen, was detected in 16 samples; C. muris was detected in only 1 sample suggesting water contamination by rodents.
Curiously, as previously mentioned, no PCR amplification was obtained from 59 samples. This problem has been described by other authors [29]. We have no clear explanation for that. Our sensitivity analysis indicates simultaneously a lack of reproducibility in the PCR analysis and the difficulty to achieve amplification in samples with low levels of contamination. It has been suggested that sufficient amounts of DNA may not be present (empty oocysts or cysts after excystation) or lack of PCR amplification may be due to the presence of inhibitors of PCR in the samples [30]. The IMS procedure applied over the samples for parasite isolation does not guarantee a complete purity of the sample. Thus, optimization of DNA extraction and amplification protocols is warranted.
The results obtained in the present study suggest that cryptosporidiosis and giardiosis should be considered very important public health issues in the north of Portugal. Also, the genetic characterization of Cryptosporidium spp. and G. duodenalis supports the possibility that there is a greater risk of infection by G. duodenalis for humans, while Cryptosporidium spp. poses a greater risk for animals. Thus, systematic monitoring of drinking water, livestock, and human biological samples are needed for risk assessment of both diseases. National Health Authorities should consider the urgent implementation of a national monitoring program for microbiological quality of drinking water that includes Cryptosporidium spp. and G. duodenalis analyses. These activities are fundamental steps to understand better the epidemiology of the infection and to allow the implementation of risk analysis models for those infections.
ACKNOWLEDGEMENTS
The authors are grateful to water-supply industry from the northern region of Portugal: Águas do Douro e Paiva, Águas do Cávado, Águas do Ave, Águas do Minho e Lima, Águas de Trásos-Montes e Alto Douro, Serviços Municipalizados de Viana do Castelo and Vimágua. This work was financially supported by funds of Project 61018, Action Environment and Health, from Calouste Gulbenkian Foundation, and funds of Project PTDC/CVT/103081/2008 from Portuguese Foundation for Science and Technology (FCT). Almeida A is supported by a fellowship from Portuguese Foundation for Science and Technology (FCT), reference SFRH/BD/22763/2005. This work was entirely developed according the Portuguese laws.

