Apical membrane antigen-1 (AMA-1) gene sequences of re-emerging Plasmodium vivax in South Korea
Article information
Abstract
Plasmodium vivax malaria re-emerged in South Korea in 1993, and epidemics continue since then. We examined genetic variation in the region encompassing the apical membrane antigen-1 (PvAMA-1) of the parasites by DNA sequencing of the 22 re-emerging P. vivax isolates. The genotype of the PvAMA-1, which was based on sequence data previously reported for the polymorphic regions, showed that two haplotypes were present at one polymorphic site. Compared with reported data, the two types, SKOR type I and type II, were similar to Chinese CH-10A and CH-05A isolates, respectively. Thus, the present study showed that two genotypes of AMA-1 genes coexist in the re-emerging Korean P. vivax.
Plasmodium vivax malaria was highly prevalent in South Korea until the 1960s-1970s, when it was controlled, and it completely disappeared after 1984 (Chai, 1999). However, it re-emerged in 1993 along the western part of the demilitarized zone (DMZ), the border with North Korea (Chai et al., 1994; Chai, 1999). Since its re-emergence, the annual number of cases has grown exponentially, and totaled 16,435 cases by the end of 2001 (National Institute of Health, 2001, 2002). By epidemiological and clinical studies, it has been suggested that the reemerging malaria in South Korea originated from North Korea (Chai, 1999; Moon and Cho, 2001), however, molecular and genetic data are required to support this suggestion.
Populations of malaria parasites exhibit significant genetic diversity (Rich et al., 2000). With regard to P. vivax, diversity has been described in terms of the disease relapse patterns, parasite morphology, biochemistry, and drug resistance. The relapse patterns of P. vivax may result from genetic variations that allow the parasites to evade the host immune responses and to adapt to the climatic extremes of the parasite's geographic distribution (Miller et al., 1994). Analysis of the diversities of merozoite surface proteins (MSP), circumsporozoite proteins (CSP), apical membrane antigens (AMA), Duffy binding proteins (DBP), and 18S rRNA genes can provide evidence upon the geographical characteristics of different strains (Qari et al., 1991, 1992; Cheng et al., 1993; Ampudia et al., 1996; Li et al., 2001).
Apical membrane antigen-1 (AMA-1) molecules are immunogenic proteins found in the rhoptry organelles of the merozoite and later on the merozoite surface at around the time of schizont rupture (Galinski and Barnwell, 1996). AMA-1 genes have been cloned and sequenced from a number of Plasmodium species, including P. falciparum, P. vivax, P. knowlesi, P. fragile, and P. chabaudi (Preiser et al., 2000). Many single base substitutions occur in the sequences of different isolates of the same species, which provides a large number of potential alleles, and makes this gene a useful marker for parasite populations (Cheng and Saul, 1994). AMA-1 molecules are also known as potential vaccine components (Preiser et al., 2000), and data are required on the extent to which immune responses to one form of the AMA-1 can protect against infections by parasites expressing other alleles.
To date, the sequence analysis of the reemerging Korean P. vivax isolates has been undertaken with regard to MSP-1, CSP, DBP, and 18S rRNA genes (Kho et al., 1999, 2001; Chai et al., 2000; Lim et al., 2000, 2001), however, no information is available on the genetic diversity of AMA-1 genes. The present study was undertaken to obtain information on the genetic characteristics of re-emerging Korean P. vivax by examining the polymorphism of AMA-1 genes.
Blood samples were collected in 1999 (n=20) and 2000 (n=2) from 22 patients diagnosed as having P. vivax by microscopy at the Seoul National University Hospital and the Paju Medical Center, South Korea (Table 1). Informed consent was obtained from all patients to the use of their blood for this study. Patients' travel histories to endemic areas were obtained by a chart review and history taking, and were used to deduce the likely origin of the infections. All of the patients had febrile illnesses when blood samples were collected. Blood parasitemia varied from less than 182 to 5,614 parasites per microliter of blood. After identification of parasites, the whole blood was either directly frozen at -80℃ or separated into packed cells and plasma, and then frozen at -80℃. The parasite DNA was extracted from each blood sample either by proteinase K digestion followed by four rounds of phenol/chloroform extraction or by using a QIAamp DNA Mini Kit®, following the manufacturer's instructions (Qiagen, Hilden, Germany). After ethanol precipitation, the DNA was redissolved in TE buffer (10 mM EDTA, 10 mM Tris-HCl, pH 8.0) and stored at -20℃ until required.

Characteristics of donors of the re-emerging Plasmodium vivax isolates in South Korea investigated and AMA-1 genotype in each isolate
DNA was amplified by the polymerase chain reaction (PCR) using the following oligonucleotide primers that allowed the amplification of the nucleotide in the polymorphic region of the PvAMA-1 gene, corresponding to base pairs (bp) 268-778 (PH-84 strain; GeneBank™ accession number, L27503); PvAF1 (5'-CCAGCTGGAAGATGTCC TGT-3', nucleotide position 268-288) and PvAR1 (5'-TAATCCGAACTTGG CGTTTC-3', nucleotide position 758-778). Primers were used at a final concentration of 0.1 µM in a 100 µl reaction mixture (10 mM Tris-HCl, pH 8.3, 50 mM KCl, 1.5 mM MgCl2, and 0.2 mM each dNTP) containing 10 µl of DNA and 2.5 units of Taq polymerase (Roche, Mannheim, Germany). Reaction mixtures underwent 32 cycles of amplification: denaturation at 95℃ for 30 sec, annealing at 65℃ for 50 sec, and extension at 72℃ for 40 sec in a DNA thermal cycler (GeneAmp® PCR System 9600; PE Applied Biosystems, Foster city, California, USA).
Amplification products were fractionated by electrophoresis on 1.2% agarose gels and stained with ethidium bromide. The PCR products were purified using a QIAEX II gel extraction kit® (Qiagen, Hilden, Germany). DNA sequencing was carried out using the dideoxy-mediated chain termination method with the same PCR primer on sequencers (Models 373A and 377, PE Applied Bio-systems, Foster city, California, USA), using the reagents and protocols supplied by PE Applied Biosystems. Gene sequences from two Korean types of P. vivax were aligned and compared with a part of PvAMA-1 gene sequence previously published. Alignment and comparison of 138 amino acid sequences (414-nucleotide region of the PvAMA-1 gene) were facilitated by using various programs of Lagergene biocomputing software package (DNASAR Inc., version 2.1, Madison, Wisconsin, USA) and CLUSTAL W program (ver. 1.81) (Thompson et al., 1994).
A single band of 511-bp, corresponding to bases 268-778 of AMA-1 gene sequence of the PH-84 isolate, was produced after amplification. No size polymorphism of PCR product was observed among isolates examined. The 22 blood samples used for the subsequent analysis showed no evidence of mixed sequences. A 414-bp region, corresponding to base pairs 322-736 of the PH-84 isolate, in each PCR product was analyzed for polymorphisms. The sequence data (Figtree et al., 2000) of the polymorphic regions showed that two genotypes, SKOR types I and II, were present in the 22 isolates. Of the 22 isolates, 9 showed the SKOR-69 genotype (SKOR (South Korean) type I) and 13 showed the SKOR-68 genotype (SKOR type II) (Table 1). When all the P. vivax AMA-1 alleles known were analyzed in this study, it was found that point mutation occurred at 13 positions. The sequences of SKOR type I (GeneBank™ accession number AF402593) and type II (AF402594) were similar to those of the Chinese CH-10A (AF171245) and CH-05A (AF171246), respectively (Figtree et al., 2000) (Fig. 1). Compared with the sequences of other isolates, the most similar isolates were the PAL-79, INDI-8, and INDO-4 (99%). In the nucleotide sequence of SKOR type I (similar to CH-10A), a G565 (K189 in amino acid) replaced A565 (E189) of SKOR type II (CH-05A). This position (565) was a fully synonymous polymorphism.
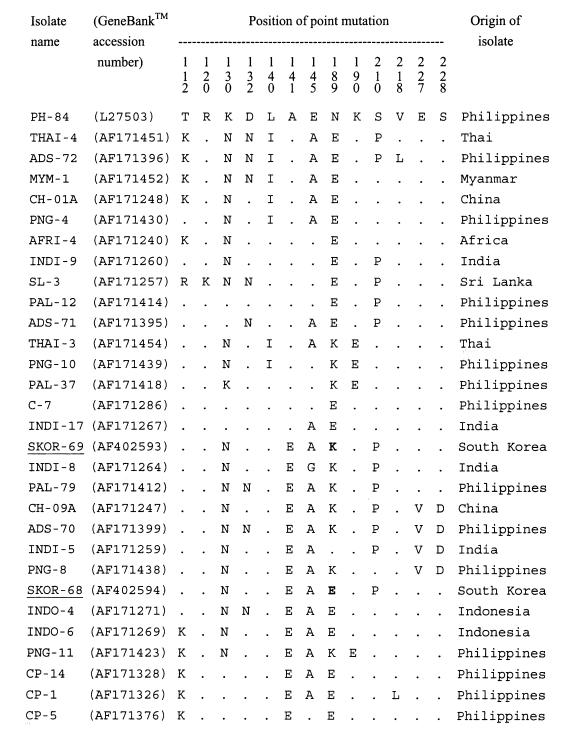
Multisequence alignment of the amino acid sequences of a number of AMA-1 proteins from various isolates of Plasmodium vivax. The alignment of sequences is based on the PH-84 sequence (Cheng and Saul, 1994). Dots indicate identical residues. Bold letters indicated dimorphic mutation site between SKOR 69 and SKOR 68 isolates.
The present results show that two genotypes of the PvAMA-1 exist among the re-emerging Korean P. vivax, and that they are similar to two Chinese genotypes. It could infer that the re-emerging malaria originated from China or another Asian country including North Korea, in which similar genotypes of the AMA-1 may exist. There had been little published information on malaria situation in North Korea. A World Health Organization Officer, recently, mentioned occurrence of about 300,000 cases in North Korea in 2001 (Moon and Cho, 2001). However, North Korean isolates of P. vivax are difficult to obtain, and could not be used in this study.
Sequence polymorphisms of AMA-1 genes provide information on the population diversity of parasites both within and between geographical areas (Figtree et al., 2000). From PvAMA-1 gene sequence analysis of 219 isolates obtained from 9 countries, 69 haplotypes have been identified at 22 polymorphic sites in the 414-bp region (Figtree et al., 2000). The polymorphism in AMA-1 genes of the re-emerging South Korean isolates was similar to two Chinese isolates, CH-05A and CH-10A. Of 7 AMA-1 haplotypes of P. vivax among Chinese isolates, only 5 were found to be unique to China (Figtree et al., 2000). The Chinese CH-05A isolates were collected during July-August 1990 from seasonal contract miners at the Hongmao coal mine located near the border of Guangxi and Guizhou Province, and the CH-10A from patients in Jiangsu and Yunnan Provinces during July-October 1993 (Figtree et al., 2000). It has been suggested that these two genotypes of the AMA-1 might be common to East Asian countries including China and South Korea.
Gene sequence analysis of the re-emerging P. vivax was performed on CSP, DBP, MSP-1 (variable block 5-6), and 18S rRNA gene, using South Korean isolates (Kho et al., 1999, 2001; Chai et al., 2000; Lim et al., 2000, 2001). Kho et al. (1999) reported that CSP genes were two haplotypes, SK-A and SK-B, and they were similar to Chinese isolates (CH-5) and the North Korean strain, respectively. They could be classified as the East Asian group (Kho et al., 1999). This Chinese CH-5 is the same isolate as that used to analyze the sequence of AMA-1 genes in the present study. Similar to the CSP, two genotypes of the DBP coexist among South Korean isolates (Kho et al., 2001). To date, the genetic study of the DBP of P. vivax has been done only in Papua New Guinea, Columbia, and South Korea, and further studies are required to understand the correlation between DBP and geographical distributions of P. vivax (Kho et al., 2001).
The present results on AMA-1 genes together with a review of literature on CSP genes strongly suggested that the recent reemergence of P. vivax in South Korea originated from East Asia. The present study is also significant, because this is the first report on PvAMA-1 genes of the re-emerging P. vivax in South Korea.
Notes
This work was supported by a grant from the Ministry of Health and Welfare, Republic of Korea(HMP-99-04-0002).