Cryptosporidium parvum is an important agent in the aetiology of the neonatal diarrhoea syndrome of calves, lambs and goat kids, causing considerable direct and indirect economic losses (
de Graaf et al., 1999a). The infection is self-limiting and immunity is based on intestinal immunoglobulins and T-cell responses (
de Graaf et al., 1999b). The
C. parvum antigens dominantly recognized by antibodies from a broad range of immune animals have been described (
Reperant et al., 1994). Screening of expression libraries with antibodies resulted in the cloning of 3
Cryptosporidium sporozoite surface antigens: CP15 (
Jenkins and Fayer, 1995), CP15/60 (
Jenkins et al., 1993) and P23 (
Perryman et al., 1996). CP15 (
Sagodira et al., 1999) and P23 (
Perryman et al., 1999) have successfully been implemented in the development of a passive vaccine against cryptosporidiosis in ruminants. In such a passive vaccinal approach the newborns are protection against cryptosporidial infection by passive transfer of hyperimmune colostrum from their immunized dams. The oral administration of anti-CP15/60 IgA monoclonal antibodies to suckling mice also provided protection against infection (
Tilley et al., 1991). Beside these sporozoite surface antigens, the micronemal proteins are likewise considered interesting target molecules for immunoprophylaxis as they too are involved in parasite invasion into host cells (
Prickett et al., 1994). This study was aimed to discover new
C. parvum sporozoite surface or micronemal antigens and to test their antigenicity in relation to humoral immunity of the bovine host.
In order to select for membrane bound (surface) or vesicle enclosed (micronemal) antigens we developed a hyperimmune rabbit serum against insoluble fragments of ultrasonicated oocysts and used it for screening a C. parvum λgt11 cDNA library. C. parvum oocysts were isolated from faeces of diseased animals by biphasic diethyl ether/PBS extraction and differential centrifugation on Percol. Cytoplasmatic compounds were released by ultrasonication and removed after centrifugation. Insoluble fragments were resuspended in PBS and emulsified with complete Freund's adjuvant for a first s.c. immunisation of Minimum Disease Level rabbits, and with incomplete Freund's adjuvant for the 2 following i.m. booster injections given at 3 and 5 weeks intervals respectively. The collected hyperimmune rabbit serum (R3αCpUnsol) recognized a complex band pattern in Western blots of insoluble oocyst fragments that were boiled in Laemmli sample buffer (not shown).
We screened a
C. parvum sporozoite and oocyst λgt11 cDNA library (
Petry et al., 1998) according to the immunological screening protocol of Sambrook et al. (
1989). The 10 clones that were recognized by R3αCpUnsol and not by pre-immune rabbit serum, were isolated after several rounds of re-screening. The inserts of 4 clones (Cp18.2.1, Cp20.2.1, Cp21.2.1 and Cp22.4.1) were amplified by PCR using the 5' and 3' LD Amplimers of the λgt11 LD-Insert Screening Amplimer Set (Clontech Laboratories, Palo Alto, CA) and sequenced by Eurogentec s.a. (Seraing, Belgium) according to the Single Run service, meaning that their DNA sequence was read only once from one of the two λgt11 primers. The sequencing data revealed that all the 4 λgt11 clones were constructed with an analogous cDNA fragment, although in 3 of them this fragment was cloned in the reversed orientation (Cp18.2.1, Cp20.2.1 and Cp21.2.1). It is not clear to us how these 3 clones could have expressed their
Cryptosporidium gene product properly. Only in the λgt11 clone Cp22.4.1 the fragment was cloned in same orientation as the λgt11 β-galactosidase gene in which it was inserted.
The amplicon of clone Cp22.4.1 was subcloned in pUC18 using Ready-To-Go T4 DNA ligase (Amersham Pharmacia Biotech Benelux, Roosendaal, Netherlands) and DH5α competent cells, and sequenced twice according to the Double Run service (Eurogentec s.a.), meaning that finally every base pair was read at least twice in each orientation. The insert of clone Cp22.4.1 had a total length of 1045 bp (excluding the flanking EcoRI adapters from the library construction) and its nucleotide sequence data are given in
Fig. 1 (also GenBank™ acc. no. AY017370). The second frame showed an open reading frame of 1004 bp. However, since the translated aa sequence that preceded the first methionine did not show any homology with the known proteins (BLASTP, National Center for Biotechnology Information;
Altschul et al., 1997), we assume that the coding region starts at this first ATG codon (assigned as position 1 in
Fig. 1) and ends at position 696 (including the stop codon TAA). This was further supported by the fact that the start methionine lays in the consensus PuNNATGPu sequence (where Pu stands for a purine and N for any base). This coding region corresponds to a protein of 231 aa with 4 zinc-finger domains characterized by a Cys-X2-Cys-X4-His-X4-Cys motif (where X can be any residue). The CCHC motif has been found mainly in the nucleocapsid protein of retroviruses where it plays a role in the packaging of the viral genomic RNA (
De Guzman et al., 1998), and also in cellular nucleic acid binding proteins which are involved in vertebrate embryogenesis (
Flink et al., 1998). Among parasitic organisms, this zinc-finger domain has been described in
Leishmania major,
Trypanosoma brucei brucei,
Trypanosoma cruzi,
Trypanosoma equiperdum and
Crithidia fasciculata (acc. no. Q04832, AJ132959, AF091396, AAB47542 and A54598 respectively). The role of two of these proteins has been reported: the HEXBP of
L. major binds to single stranded DNA containing a hexanucleotide repeat (
Webb and McMaster, 1993) and the universal minicircle sequence binding protein of
C. fasciculata binds to the conserved universal sequence of kinetoplast DNA minicircles (
Abeliovich et al., 1993). It should be noted that another
C. parvum zinc-finger sequence has been described (acc. no. U48717). Further, the EST and GST projects have yielded at least 5 putative zinc-finger sequences: CpEST.387, CpEST.088, CpEST. 197, CpG0020B and CpG0306A (more information on the world wide web
http://www.ebi.ac.uk/parasites/cparv.html).
In order to study the antigenicity of the Cp22.4.1 protein, the coding region of the clone (stop codon excluded) was subcloned in a pBAD-TOPO-TA expression vector (Invitrogen, Carlsbad, CA). We designed two primers for the amplification of the corresponding fragment by PCR: forward primer 5'-ATGATAAATGAAAGCATTCAAAA-3' and reverse primer 5'-CATTGCTTCATTCCACATTAA-3'. We have chosen to use sporozoite DNA as template for this PCR reaction, as it would allow us to confirm the origin of the cloned gene (=
C. parvum) and to compare its DNA sequences at the pre-transcriptional (chromosomal DNA) and post-transcriptional (cDNA) level. Cloning of the PCR-product in pBAD-TOPO-TA was according to manufacturer's instructions and the resulting pBAD clone was proven to have an insert that was 100% identical to that of the Cp22.4.1 λgt11 clone. Thus, the chromosomal organisation of this gene lacks introns. This sequencing result permitted us to use the name Cp22.4.1 also for the pBAD clone and its recombinant expression product (= His-tagged Cp22.4.1 protein). Optimal expression was obtained by stimulating liquid cultures at OD
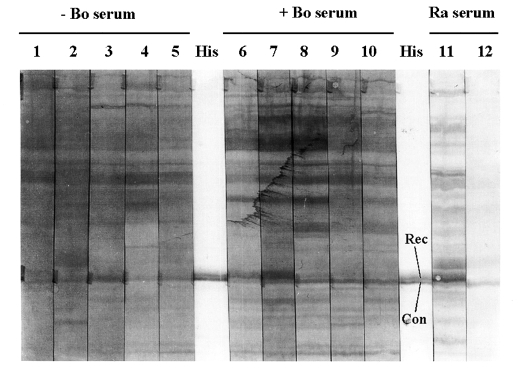
600 = 0.5 with 0.002% L-arabinose (or more) and by harvesting cells after 4 h incubation at 37℃. These cells boiled in Laemmli sample buffer were used to test bovine humoral recognition of the His-tagged Cp22.4.1 protein on Western blots. An anti-His (C-terminal)-HRP antibody served to localize Cp22.4.1 protein on the blots, but revealed a double band of approximately 35 kDa (
Fig. 2). Comparison with bacterial lysates from other His-tagged
Cryptosporidium antigens (not shown), and staining with the rabbit serum R3αCpUnsol (
Fig. 2), revealed that only the upper band corresponded with the His-tagged Cp22.4.1 protein. At the same altitude a band was recognized by 2 out of 5 sera from calves that recovered from natural
Cryptosporidium infection. This band was not visualized using sera from neonatal animals (aged between 8 and 10 days) that were kept free of infection by Halocur (Intervet Belgium) treatment (
Fig. 2).
In conclusion, our efforts to clone new surface and micronemal antigens resulted in the finding of a λgt11 clone corresponding to a protein with 4 zinc-finger domains that was recognized by serum antibodies from immune neonatal calves in a specific way. It would be interesting to study the cellular localization (most probably nuclear) and function (most probably in gene regulation) of this protein in further projects.
Notes
-
This work was supported by a grant from the Belgian Ministry of Small Enterprises, Traders and Agriculture (Dienst Landbouwkundig Onderzoek; S-5840).
References
- 1. Abeliovich H, Tzfati Y, Shlomai J. A trypanosomal CCHC-type zinc finger protein which binds the conserved universal sequence of kinetoplast DNA minicircles: isolation and analysis of the complete cDNA from Crithidia fasciculata. Mol Cell Biol 1993;13:7766-7773.
- 2. Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997;25:3389-3402.
- 3. de Graaf DC, Vanopdenbosch E, Ortega-Mora LM, Abbassi H, Peeters JE. A review of the importance of cryptosporidiosis in farm animals. Int J Parasitol 1999a;29:1269-1287.
- 4. de Graaf DC, Spano F, Petry F, Sagodira S, Bonnin A. Speculation on whether a vaccine against cryptosporidiosis is a reality or fantasy. Int J Parasitol 1999b;29:1289-1306.
- 5. De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 1998;279:384-388.
- 6. Flink IL, Blitz I, Morkin E. Characterization of cellular nucleic acid binding protein from Xenopus laevis: expression in all three germ layers during early development. Dev Dyn 1998;211:123-130.
- 7. Jenkins MC, Fayer R, Tilley M, Upton SJ. Cloning and expression of a cDNA encoding epitopes shared by 15- and 60-kilodalton proteins of Cryptosporidium parvum sporozoites. Infect Immun 1993;61:2377-2382.
- 8. Jenkins MC, Fayer R. Cloning and expression of cDNA encoding an antigenic Cryptosporidium parvum protein. Mol Biochem Parasitol 1995;71:149-152.
- 9. Perryman LE, Jasmer DP, Riggs MW, Bohnet SG, McGuire TC, Arrowood MJ. A cloned gene of Cryptosporidium parvum encodes neutralization-sensitive epitopes. Mol Biochem Parasitol 1996;80:137-147.
- 10. Perryman LE, Kapil SJ, Jones ML, Hunt EL. Protection of calves against cryptosporidiosis with immune bovine colostrum induced by a Cryptosporidium parvum recombinant protein. Vaccine 1999;17:2142-2149.
- 11. Petry F, Shirley MW, Miles MA, McDonald V. Characterisation of a Cryptosporidium parvum-specific cDNA clone and detection of parasite DNA in mucosal scrapings of infected mice. Mol Biochem Parasitol 1998;95:21-31.
- 12. Prickett MD, Smarz TR, Adams JH. Dimorphism and intergenic recombination within the microneme protein (MP-1) gene family of Plasmodium knowlesi. Mol Biochem Parasitol 1994;63:37-48.
- 13. Reperant JM, Naciri M, Iochmann S, Tilley M, Bout DT. Major antigens of Cryptosporidium parvum recognised by serum antibodies from different infected animal species and man. Vet Parasitol 1994;55:1-13.
- 14. Sagodira S, Buzoni-Gatel D, Iochmann S, Naciri M, Bout D. Protection of kids against Cryptosporidium parvum infection after immunization of dams with CP15-DNA. Vaccine 1999;17:2346-2355.
- 15. Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 1989, Cold Spring Harbor, USA. Cold Spring Harbor Laboratory Press.
- 16. Tilley M, Upton SJ, Fayer R, et al. Identification of a 15-kilodalton surface glycoprotein on sporozoites of Cryptosporidium parvum. Infect Immun 1991;59:1002-1007.
- 17. Webb JR, McMaster WR. Molecular cloning and expression of a Leishmania major gene encoding a single-stranded DNA-binding protein containing nine "CCHC" zinc finger motifs. J Biol Chem 1993;268:13994-14002.
Fig. 1Complete nucleotide sequence data of the λgt11 clone Cp22.4.1 ending with a 17-mer poly-A tail. The deduced aa sequence is given from the first ATG-codon (in bold and underlined) to the first TAA-stop codon at position 694-696 (also in bold and underlined). The zinc-finger domains characterized by a Cys-X2-Cys-X4-His-X4-Cys motif are marked in gray.

Fig. 2Bovine immune response against the Cp22.4.1 protein. Western blots of the bacterial lysate of the pBAD clone that expressed the His-tagged Cryptosporidium protein. An anti-His (C-terminal)-HRP antibody (His), together with immune (= R3αCpUnsol; lane 11) and pre-immune (lane 12) rabbit serum (Ra serum), served to localize the Cp22.4.1 protein on the blots (Rec = recombinant Cp22.4.1 protein; Con = immuno-recognized contaminant). At the same altitude as Cp22.4.1 (approximately 35 kDa; molecular mass was determined by introducing markers on an other - not shown - blot) a band was visualized by 2 (lanes 6 and 7) out of 5 sera from Cryptosporidium immune animals (+ Bo serum; lanes 6-10) and not using sera from parasite naive animals (- Bo serum; lanes 1-5). The secondary antibody was an anti-bovine IgG (whole molecule) peroxidase conjugate.

Citations
Citations to this article as recorded by

-
Evaluation of Recombinant Oocyst Protein CP41 for Detection of
Cryptosporidium-
Specific Antibodies
Sonia A. Kjos, Mark Jenkins, Pablo C. Okhuysen, Cynthia L. Chappell
Clinical and Vaccine Immunology.2005; 12(2): 268. CrossRef