Identification of newly isolated Babesia parasites from cattle in Korea by using the Bo-RBC-SCID mice
Article information
Abstract
Attempts were made to isolate and identify Korean bovine Babesia parasite. Blood samples were collected from Holstein cows in Korea, and Babesia parasites were propagated in SCID mice with circulating bovine red blood cells for isolation. The isolate was then antigenically and genotypically compared with several Japanese isolates. The Korean parasite was found to be nearly identical to the Oshima strain isolated from Japanese cattle, which was recently designated as Babesia ovata oshimensis n. var. Haemaphysalis longicornis was the most probable tick species that transmited the parasite.
INTRODUCTION
Babesiosis is a tick-borne disease of domestic and wild mammals caused by protozoan parasites of the genus Babesia. The acute disease is occasionally seen in cattle, which is characterized by fever, hemolytic anemia, icterus, hemoglobinuria and death. However, chronic infections with inapparent clinical symptoms also occur frequently in cattle and other animals as well. In such cases, the parasite infection persists for a long time and decreases animal productivity. Moreover, when chronic infection is combined with other factors, such as other infections, stress, shipping, and delivery, the disease may become severe and can result in fatal outcome (Soulsby, 1982).
It has been known since 1912 that piroplasms were parasitized in Korean cattle. In a study on the classification and distribution of ticks among Korean cattle, it was found that Boophilus microplus and Haemaphysalis longicornis were the predominant species (Han et al., 1966). Lee and Choi (1976) reported the first unequivocal observation on bovine babesiosis in Korean native cattle. They described that the intraerythrocytic protozoa were somewhat different from the large type Piroplasma; the organisms predominantly appeared as a single ring form rather than paired pyriform. Isolation of a Babesia parasite from Korean cattle was carried out by Jeon in 1978; he compared it with B. bigemina and B. ovata, and concluded that the organism was very similar, if not identical, to the Miyake strain of B. ovata from Japan (Minami and Ishihara, 1980). Suh (1987) did much closer examinations on the Korean bovine Babesia parasite; he concluded that it was identical to B. ovata isolated from Japanese cattle. However, conclusions of those studies were based primarily on morphological and serological characteristics of the parasites, and detailed analyses with modern molecular biological techniques have not yet been carried out.
One of the major difficulties associated with studies of intraerythrocytic protozoa has been the unavailability of appropriate in vivo experimental systems. Taking advantage of the characteristics of severe combined immune deficiency (SCID) mouse (Bosma et al., 1983; McCune et al., 1988), Tsuji et al. (1992) were able to produce the mice with circulating bovine erythrocytes (Bo-RBC-SCID mice) in which Theileria sergenti proliferated. Later, they used this SCID mouse model for isolation of a Babesia parasite from grazing calves in Japan (Tsuji et al., 1995). Although the initial blood samples that they collected from calves had both Babesia and Theileria parasites, they were able to isolate only the Babesia parasites by repeated passages in mouse model as they grew much more rapidly than Theileria.
In the present study, attempts were made to isolate and identify Babesia parasites from cattle in Korea. The intraerythrocytic protozoan parasites were propagated in Bo-RBC-SCID mice, and compared morphologically, antigenically and genotypically with several bovine Babesia sp. In order to identify a possible tick vector, we also collected ticks from cattle and vegetation where parasites were isolated.
MATERIALS AND METHODS
Collection of Korean Babesia sp.
Three farms were selected for parasite collection. Farm A and B were located in Chungnam Province and Farm C in Jeonnam Province. Blood samples were taken from 20 adult cattle of each farm; blood smears were prepared and then stained with Giemsa's stain. Five Holstein cows of Farm C only were found infected with Babesia, and 5 ml of blood were taken from each cow for later use.
SCID mice
An in vivo experimental system using SCID mice for Babesia species has been previously described (Tsuji et al., 1995). Briefly, C.B-17 scid/scid mice (Japan CLEA, Tokyo, Japan) of mixed sex and between 7 and 14 weeks old were used. The mice were housed in a vinyl-film isolator at around 23℃ and provided with λ-ray-irradiated pellets (FR-1; Funabashi Co., Japan) and autoclaved tap water ad libitum. All mice were splenectomized under anesthesia and were used for experiments after the surgical wounds had healed completely. They were treated according to Laboratory Animal Control Guidelines at Rakuno Gakuen University, which basically conform to the American Association of Laboratory Animal Control Guidelines issued by the National Institute of Health.
Bo-RBC-SCID mice
The SCID mice with circulating bovine red blood cells were prepared as described previously (Tsuji et al., 1995). To achieve the rapid substitution of circulating RBCs in SCID mice with Bo-RBCs, the authors developed a rat-mouse hybridoma, clone 2E11, producing anti-mouse RBC monoclonal antibody. The cultured supernatants of this hybridoma were concentrated into 1/20 of original volume by ammonium sulfate precipitation followed by dialysis against 0.85% saline. Splenectomized SCID mice were periodically administered subcutaneously with 200 ul of concentrated supernatants on Days -2, 1, and 5 in addition to intravenous transfusion of 0.5 ml packed cell volume of parasite-free Bo-RBCs.
Isolation of Korean Babesia parasites
The blood samples from two Holstein cows, which contained relatively high numbers of Babesia-infected RBCs, were chosen for parasite isolation. The RBC samples were washed three times with 0.85% saline, and 0.5 ml of packed cells was intravenously inoculated into a Bo-RBC-SCID mouse. Two mice were used for each sample. 10-20µl of tail blood samples of the mice were collected in heparinized tubes every day. The percentages of Bo-RBCs and parasitized Bo-RBCs in the peripheral blood cells was determined by Giemsa staining for blood smears and flow cytometry (Cyto ACE-150, JASCO Co., Japan) for RBC samples stained with FITC-labeled anti-Bo-RBC antibody, respectively (Tsuji et al., 1992). Babesia preparations that were free from Theileria were obtained by successive passages of the Babesia-infected RBCs in Bo-RBC-SCID mice as previously described (Tsuji et al., 1995; Terada et al., 1995). Babesia-infected Bo-RBCs obtained from the infected Bo-RBC-SCID mice were either passaged into new, uninfected Bo-RBC-SCID mice or suspended in a cell freezing solution (Cell Banker, Nippon Zenyaku Co. Ltd., Japan) followed by cryopreservation in liquid nitrogen. When needed, the frozen parasites were propagated by intraperitoneal inoculation into Bo-RBC-SCID mice. The paired, pyriform Babesia sp. in blood smears stained with Giemsa was measured, together with other B. ovata (strains Oshima and Miyake), B. bigemina (strain GJ29) and B. bovis (strain Mo7), in length and width with a real-time image analyzer (LUZEX F, Nikon, Japan).
Antigenic analysis
Western blot analysis was performed as described (Arai et al., 1998). Frozen stocks of Babesia-infected RBC were thawed and washed five times with 10 mM Tris-HCl containing 10 mM EDTA (pH 7.5) at 4℃ and then centrifuged at 10,000g for 10 min. The resulting pellets were resuspended in 125 mM Tris-HCl (pH 6.5) containing 5% mercaptoethanol, 2% sodium dodecyl sulfate, 10% glycerol, and 0.1% bromophenol blue, heated at 98℃ for 5 min, and vigorously vortexed.
The samples were diluted such that contained material from equivalent numbers of infected RBC and were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE; Laemmli, 1970) with 7.5% acrylamide gels, together with B. ovata (Oshima, Miyake and Okushiri strains), B. bigemina and B. bovis. Proteins were electrophoretically transferred to Fluorotrans membranes (Pall BioSupport, USA) for 1 h at 10 V. After blocking with PBS containing 0.5% casein, membranes were reacted with appropriately diluted immune sera, followed by reaction with secondary antibodies (alkaline phosphatase-conjugated Affinipure goat anti-mouse IgG (Organon Teknika, UK). Immunoreactive antigens were detected with a BCIP/NBT Alkaline Phosphatase Substrates Kit IV (Vector Laboratories, Inc., USA).
DNA preparations and sequence analysis
DNA samples were prepared from blood samples with a whole blood DNA extraction kit (GenTLE, TaKaRa Biochemical, Japan). Sequences encoding eukaryotic small-subunit rRNA (rDNA) were amplified from the DNA samples by PCR with primer set described by Medlin et al. (1988).
PCR mixtures contained 400 mM each deoxynucleoside triphosphates, 0.25 mM each primer (rRNA-3': 5'-GTCTTAGTATAAGCTTTTATACAGCG-3'; rRNA-5': 5'-GATAGGTCAGAAACTTGAATGATACATCG-3'), 10 to 100 ng of template DNA, and 2.5 U of La Taq DNA polymerase (TaKaRa Biochemical, Japan) in 50 ul of PCR buffer supplied together with enzyme. Thermal cycling was carried out in a Gene-Amp PCR system 9600 thermal cycler (Perkin-Elmer, USA), with 30 cycles of denaturation at 94℃ for 2 min, annealing at 55℃ for 2 min and extension at 72℃ for 90 sec.
The specific PCR products (1.7 kb) were purified by agarose gel electrophoresis, followed by cloning into EcoRV site of pT7blue T-vector (Novagen, Germany) according to the manufacturer's instruction. Nucleotide sequences were determined by the dideoxynucleotide chain termination method (Sanger et al., 1977) with double-stranded plasmid DNA as a template. Sequencing reactions were carried out with an AutoRead DNA sequencing kit (Pharmacia, Sweden) with fluorescein isothiocyanate-labeled primers. Samples were analyzed with an ALF DNA Sequencer II sequencer (Pharmacia, Sweden), and sequence data were processed with the associated software (ALF manager, version 3.02). The sequences of both strands of the rRNA gene (rDNA) were determined and were submitted to GenBank.
Phylogenetic analysis
Analyses of DNA sequences and phylogenetic relationships were done by using the MacVector software package, version 7.0 (Genetic Computer Group Inc., USA). The rDNA sequences (accession numbers are given in parentheses) for phylogenetic analysis were from B. bigemina (X59604), B. divergens (U16370), B. canis (L19079), B. caballi (Z15104) B. equi (Z15105), B. rodhaini (AB04999), B. microti (U09833) and B. gibsoni (L13729). The sequences were aligned with Program Clustal W Alignment (Higgins et al., 1992), and a phylogenetic tree by neighbor-joining method (Saitou and Nei, 1987) was constructed from the aligned sequences by the programne Phylogenetic Analysis in the MacVector software. Support for tree nodes was calculated by 1,000 bootstrap replicates by the Bootstrap Tree algorithm.
Collection and identification of vector ticks
The vector ticks of Korean Babesia were collected from 5 Holstein cows, which were infected with Babesia parasites. Twenty-one adult ticks were collected from the neck and ears of the cow; 15 young ticks were collected by flagging vegetation around pastureland. All ticks were sent to the Protozoology Section of the National Institute of Infectious Diseases in Japan for identification.
RESULTS
The morphological feature of Korean Babesia sp. was typical paired pyriform or oval (Fig. 1). The size of Korean Babesia sp. merozoite was summarized together with those of four other Babesia sp. (Table 1). The western blot analysis performed with five other Babesia sp. clearly showed that Korean Babesia parasite closely resembled the Oshima strain of B. ovata (Fig. 2).

Photomicrograph of Korean Babesia species propagated in Bo-RBC-SCID mouse. The Giemsa-stained thin-smear blood film was prepared 19 days after inoculation of the bovine blood specimen, when bovine RBCs made up 90% of the peripheral RBCs of the mouse.

The measurement of Korean Babesia sp. merozoites compared with Japanese B. ovata (Oshima and Miyake strains), B. bigemina and B. bovis

SDS-PAGE analysis of Korean Babesia sp. together with Japanese B. ovata (Oshima, Miyake and Okushiri strains), B. bigemina and B. bovis.
Parasite DNAs were prepared from blood samples of the Bo-RBC-SCID mice, and the rDNA were amplified and sequenced. A computer search of the sequences in GenBank showed the highest degree of sequence homology with the rDNA of the Oshima strain of B. ovata, although differences were seen at 26 positions in the 1,695 bp sequence. The sequence of PCR products of the newly isolated Korean bovine Babesia sp. was 1,695bp in length and showed 95% sequence homology to the SSU rRNA gene from other Babesia parasites, Oshima and Okushiri strains of B. ovata, and B. bigemina (Fig. 3).

Sequences of the SSU-rRNA genes of Korean Babesia sp. (KBS), B. ovata oshima (BOO), B. bigemina (BBI), and B. ovata okuma (BOK). Gaps (-) represent spaces introduced into the aligned sequences by the Multiple Alignment program in the CLUSTAL V program.
The phylogenetic relationships among Korean Babesia sp., other Babesia sp. and Theileria sp. were clearly indicated that Korean Babesia sp. is the most closely related to B. ovata Oshima (Fig. 4).
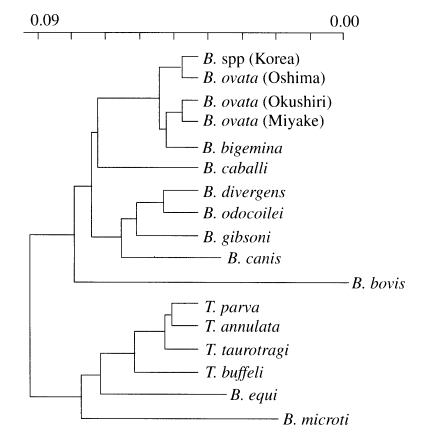
Phylogenetic tree constructed with rDNA sequences of various apicomplexan parasites such as Babesia and Theileria sp.
All ticks collected from both cattle and vegetation were morphologically identified as Haemaphysalis longicornis (Fig. 5), which appeared to serve as the vector for the Korean bovine Babesia parasite.
DISCUSSION
In the present study, we attempted to identify the Babesia sp. isolated in Korea, which is known since 1912 to inhabit Korean cattle (Han et al., 1966), using recent molecular biological techniques. The vector ticks for parasite were collected from cattle and vegetation where the parasite was present and identified.
The Korean Babesia sp. was purified by the methods described previously (Terada et al., 1995; Tsuji et al., 1995), and it was then compared with several Japanese Babesia sp. morphologically, immunologically and genetically. The results of the present study clearly indicated that the Korean Babesia sp. is very close to B. ovata oshimensis in Japan (Ohta et al., 1996). Previous reports concluded that the Korean Babesia sp. was either very similar (Jeon, 1978) or identical (Suh, 1987) to Japanese B. ovata Miyake strain (Minami and Ishihara, 1980).
The B. ovata oshimensis, which was newly isolated in 1993 from Japanese Brown cattle in Oshima, Hokkaido and temporarily named as Babesia sp. 1, was somewhat different from the Miyake strain, the type strain of B. ovata isolated in 1980 (Minami and Ishihara, 1980; Ohta et al., 1995). After transmissibility of the parasite with vector ticks, it was named B. ovata oshimensis n. var. (Ohta et al., 1996).
Various species of Ixodid ticks have been identified as vectors and reservoirs of important viral, rickettsial and protozoal pathogens of man and other animals. A large number of species occur in the genus Haemaphysalis. Of those, H. longicornis has a wide distribution throughout the East, occurring in China, Japan, Australia and New Zealand, and inhabits man, cattle, sheep, horse and dog. Heavy infections may be seen in cattle (Soulsby, 1982). It was found that the large type Babesia was transmitted by H. longicornis (Minami and Ishihara, 1980; Ohta et al., 1996). The ticks collected in the present study were identified as H. longicornis (Fig. 5), indicating that it plays a major role in transmitting bovine Babesia parasites in both Korea and Japan.
Notes
This study was supported by Korea-Japan Researchers' Friendship Programme Grants of Korea Science and Engineering Foundation (KOSEF) supported this work.
