Isolation and characterization of a cDNA encoding a mammalian cathepsin L-like cysteine proteinase from Acanthamoeba healyi
Article information
Abstract
We have cloned a cDNA encoding a cysteine proteinase of the Acanthamoeba healyi OC-3A strain isolated from the brain of a granulomatous amoebic encephalitis patient. A DNA probe for an A. healyi cDNA library screening was amplified by PCR using degenerate oligonucleotide primers designed on the basis of conserved amino acids franking the active sites of cysteine and asparagine residues that are conserved in the eukaryotic cysteine proteinases. Cysteine proteinase gene of A. healyi (AhCP1) was composed of 330 amino acids with signal sequence, a proposed pro-domain and a predicted active site made up of the catalytic residues, Cys25, His159, and Asn175. Deduced amino acid sequence analysis indicates that AhCP1 belong to ERFNIN subfamily of C1 peptidases. By Northern blot analysis, no direct correlation was observed between AhCP1 mRNA expression and virulence of Acanthamoeba, but the gene was expressed at higher level in amoebae isolated from soil than amoeba from clinical samples. These findings raise the possibility that Ahcp1 protein may play a role in protein metabolism and digestion of phagocytosed bacteria or host tissue debris rather than in invasion of amoebae into host tissue.
INTRODUCTION
Members of the genus Acanthamoeba, one of the amphizoic amoebae with ubiquitous distribution, can cause sight-threatening keratitis and life-threatening granulomatous amoebic encephalitis (GAE) (Marciano-Cabral et al., 2000). Considering the high phagocytic activity of Acanthamoeba as a free-living organism and tissue invasive behavior as a parasite, it can be speculated that proteinases should be involved with many processes in the amoeba. Lehr et al., (1998) found that mannose induced cytopathic effect of Acanthamoeba culture supernatant was inhibited by PMSF, a serine proteinase inhibitor. Kong et al. (2000) reported strong proteinase activity of Acanthamoeba from culture supernatant and amoeba lysate. They also proposed that the purified serine proteinase from culture supernatant would play a role in host tissue invasion because it had strong proteolytic activity against ECM proteins like type I and IV collagens and fibronectin. Additionally, a cDNA that encoded subtilisin like serine proteinase of A. healyi was also identified and characterized (Hong et al., 2000).
In other protozoan parasites, cysteine proteinases have been known to play important roles in the metabolism, development, or survival of protozoa. For example, cysteine proteinases of Plasmodium spp. have been shown to degrade host hemoglobin or to cleave ankyrin of erythrocyte membrane to facilitate parasite release (Rosenthal et al., 1988; Raphael et al., 2000). Cysteine proteinases of Trypanosomatid have been reported to do roles in developmental processes, pathogenesis and immune modulation (Tomas et al., 1997; Mottram et al., 1998). The genus Giardia has a cysteine proteinase that plays essential role in excystation (Ward et al., 1997). In the case of Entamoeba histolytica, a lot of evidences to elucidate the role of cysteine proteinase as a pathogenic factor have been reported (Que and Reed, 2000). However, some papers proposed different biological function of cysteine proteinase rather than a pathogenic factor in E. histolytica (Ankri et al., 1998). Trophozoite of which cysteine proteinase activity was strongly inhibited with antisense RNA showed a significantly lower phagocytic activity but had normal growth rate and similar cytopathic and hemolytic activity to the control (Ankri et al., 1998).
However, the identification and characterization of cysteine proteinase genes of Acanthamoeba have seldom been studied (Yun et al., 1999). In this paper, we have isolated and characterized a cDNA clone encoding a cysteine proteinase of A. healyi and described it as an orthologous cysteine proteinase to mammalian cathepsin L.
MATERIALS AND METHODS
Amoeba cultivation
Acanthamoeba healyi, originally isolated from the brain of a GAE patient, was obtained from ATCC (ATCC # 30866), and grown axenically in peptone-yeast extract-glucose (PYG) medium at 25℃. Amoebae of approx. 4.0×106 cells/ml were centrifuged at 1100 rpm for 10 min at room temperature. The cell pellet was subjected to extraction of RNA and DNA for further study.
Construction and screening of A. healyi cDNA library
Acanthamoeba healyi mRNA was prepared from trophozoites using the mRNA isolation kit (Qiagen, Germany) following the manufacturer's instruction. cDNA was synthesized using a ZAP-cDNA synthesis kit (Stratagene, California, U.S.A.). Following EcoR I adaptor ligation to the 5' terminus and Xho I digestion, the cDNA was inserted into the EcoR I-Xho I site of lambda ZAP (Stratagene, California, U.S.A.). Degenerate oligonucleotide primers for reverse transcription-PCR of A. healyi gene encoding cysteine proteinase were CPP5'(5'-ACAGAATTCCARTGYGGITCITGG-3') and CPP 3'(5'-TTAAAGCTTCCATTYTTIACRATCCARTA-3'), based on the amino acid sequences of the active sites conserved in C1 family of cysteine proteinases. Amplification reaction was 30 cycles of denaturation at 94℃ for 1min, annealing at 55℃ for 1 min, and extension at 72℃ for 1min, and the final extension step was 5 min at 72℃ in a DNA Thermal Cycler (Model 2400, Perkin-Elmer Cetus). The amplified DNA fragment was gel-purified and cloned into the Sma I-digested pBluescript SK(-) vector (Stratagene, California, U.S.A.). Recombinant phages were spread on agar plates and lifted to nylon membranes in duplicate. Membranes were hybridized to the cDNA insert of AhCP1, which had been labeled with [α-32P]dCTP using random primed DNA labeling kit (Boehringer Mannheim, Germany). The library was screened by standard methods. Co-infection with Exassist helper phage was used to rescue pBluescript phagemid from plaque-purified phage according to the manufacturer's instructions. All DNA sequencing was performed by the dideoxynucleotide method using custom synthesized primers.
Sequence analysis of AhCP1 and construction of the phylogenetic tree
Homology search was performed with the Basic Local Alignment Search Tool (BLAST) program of the National Center for Biotechnology Information, National Institutes for Health (Altschul et al., 1990; Altschul et al., 1994). The cleavage site of the AhCP1 signal peptide was predicted following Nielson et al. (1997). Phylogeny was obtained from the alignment of 26 cysteine proteinases using CLUSTALW 1.7 program with a low gap penalty using the European Bioinformatics Institute server (Pearson and Lipman, 1988). The sequence alignment of catalytic domains of cysteine proteinases was performed with the corresponding regions of various organisms (Siezen and Leunissen, 1997).
Northern blot analysis
Total RNA from A. healyi OC-3A, Acanthamoeba sp. KA/E1 and KA/E2, two corneal isolates, KA/L5 and KA/S8, two environmental isolates, A. castellanii Castellani, A. polyphaga, A. castellanii Ma, and A. mauritaniensis was extracted. Poly(A)+ RNA was denatured and separated on an 1.1% agarose gel containing 6.7% formaldehyde, and transferred to a Hybond-N+ nylon membrane (Amersham, U.S.A.). Oligonucleotide primers for PCR amplification of the catalytic domain of cysteine proteinase were CYS (5'-CAGGGTCAGTGCGGCTCGTGCTGGTCGTTCTCGACC-3') and HIS (5'-GCCCCAGCCGACGACGAGCACACCGTGGTCGAG-3'), based on the amino acid sequences Gln132 to Thr143 and Leu275 to Gly285 of AhCP1. The 462 bp PCR product of AhCP1 labeled by the random prime labeling method (Boehringer Mannheim, Germany) was used as a probe in standard Northern hybridization analysis (Sambrook and Russell, 2001)
RESULTS
PCR amplification and molecular cloning of a cysteine proteinase cDNA gene
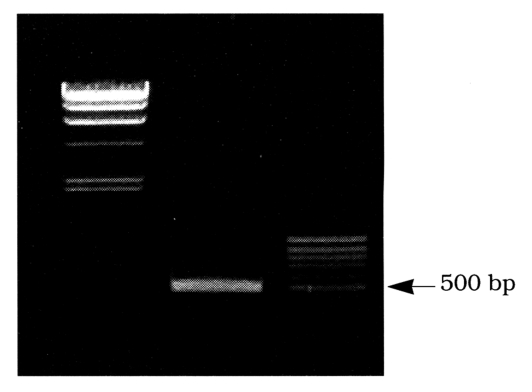
To generate a cDNA fragment of Acanthamoeba cysteine proteinase, RT-PCR using degenerate primers specific for cysteine proteinases was carried out with mRNA of A. healyi. The oligonucleotide primers were designed based on the amino acid sequences of cysteine proteinase. PCR primers, CPP5' (sense) and CPP3'(antisense) were based on the conserved motifs flanking the catalytic cysteine and asparagine residues Gln19 to Trp26 and Tyr170 to Trp177 respectively of the papain superfamily (C1) of cysteine proteinase. The PCR reaction using degenerate primers yields a cDNA fragment of approximately 500 bp (Fig. 1). The cDNA was sequenced and a comparison of the deduced amino acid sequence with the protein databases indicated strong homology with a number of cathepsin L like cysteine proteinases. To generate the full length cDNA sequence of cysteine proteinase, the cDNA fragment was used as probe for screening the A. healyi cDNA library. After three rounds of stringent hybridization, six potentially positive clones were selected. When the insert DNAs were hybridized with the cDNA fragment by Southern blot analysis, all cDNA clones hybridized to the probe and were confirmed to be the products of a single gene by sequence analysis. The largest insert had a single open reading frame of 990 bp, predicting a protein of 330 amino acid residues (Fig. 2).

Nucleotide sequence of AhCP1 derived from cDNA. The deduced amino acid sequence is shown in single letter code below the nucleotide sequence. The putative signal sequence followed by a predicted pro-region is marked by single downward arrow. The predicted end of pro-region is marked with two downward arrows. Shaded amino acids indicate the ERFNIN motif. The degenerate priming sites for RT-PCR are underlined. Residues of the cysteine proteinase catalytic triad, Cys25, His159, and Asn175 are shown in the bold face type.
Deduced amino acid sequence analysis of AhCP1
Based on homology searches using BLASTP, the deduced amino acid sequence of the cloned gene clearly belongs to the C1 peptidase (papain superfamily) proteinase clan as defined by Rawlings and Barrett (1994). Thus, the cloned gene was termed as Acanthamoeba healyi cysteine proteinase (AhCP1). When the amino acid sequence was aligned with C1 peptidase previously reported from eukaryotes or prokaryotes, AhCP1 was homologous at the core catalytic triad residues, Cys25, His159, and Asn175, cysteine proteinase defining sequence domains (Fig. 3). The active site of AhCP1 has great homology with those cysteine proteinase; 73% similarity with cysteine proteinase of Sarcophaga. Among mammalian cathepsin L and S, it displays 70% similarity with boar cathepsin L, 69% similarity with cattle, 73% with mouse and 71% with human cathepsin L, and 70% with cathepsin S. Phylogenetic analysis in this study suggested that AhCP1 belongs to the cathepsin L subgroup of the papain superfamily (Fig. 4). Sequence comparisons with other cathepsin L proteinase also suggested that Ahcp1, like majority of cysteine proteinase, could be formed as a pre-pro-protein precursor. The pre-region cleavage site of the signal sequence was predicted to be between Ala20 and Ala21 (Fig. 2, one downward arrow) by weight matrix analysis (Nielson et al., 1997). The pro-region cleavage site was predicted between Asn92 and Phe93 (Fig. 2, double downward arrows). Cleavage of pre-pro-region would reduce the molecular mass of Ahcp1 from a predicted 330 amino acid protein with a molecular mass of 36 kDa to a 216 amino acid mature protein with a molecular mass of 24 kDa. As in the other proteinases of C1 family, there was a motif GCNGG (residues 64-68) which is suggested to have an important structural role. The enzyme has neither N-terminal nor C-terminal extension observed in Plasmodium falciparum and Leishmania major cysteine proteinases. AhCP1 had a highly conserved interspersed amino acid motif, the ERFNIN motif (EX3RX2(V/I) FX2NX3IX3N) in the propeptide region (Fig. 2 & 3).
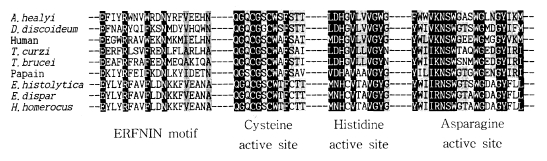
Sequence alignment of ERFNIN motif and cysteine proteinase catalytic domains of A. healyi OC-3A and 8 cysteine proteinases from other organisms. The shaded regions represent conserved amino acid sequences.
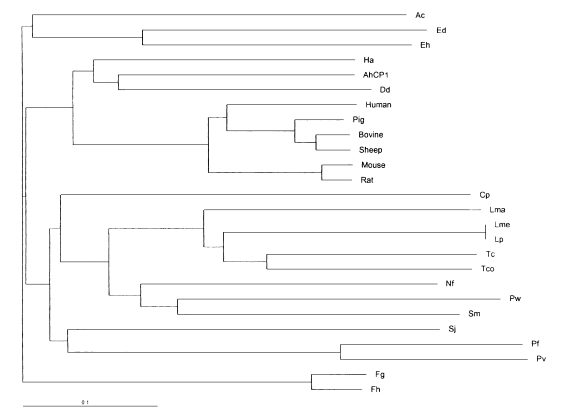
Phylogenetic tree analysis based on the sequence alignment of catalytic domains of cysteine proteinases from various organisms. Ac; Acanthamoeba culbertsoni cysteine proteinase AcCP2 (accession No. AAD02173.3), Ed; Entamoeba dispar cysteine proteinase (accession No. CAA62834.1), Eh; Entamoeba histolytica cysteine proteinase 2 precursor (accession No. Q01958), Ha; Homarus americanus digestive cysteine proteinase 1 precursor (accession No. P13277), AhCP1; Acanthamoeba healyi AhCP1, Dd; Dictyostelium discoideum cysteine proteinase 5 precursor (accession No. P54640), Human; Human cathepsin L precursor (accession No. P07711), Pig; Pig cathepsin L precursor (accession No. Q28944), Bovine; Bos taurus Bovin cathepsin L precursor (accession No. P25975), Sheep; Sheep cathepsin L (accession No. Q10991), Mouse; Mouse cathepsin L precursor (accession No. P06797), Rat; Rat cathepsin L precursor (accession No. P07154), Cp; Carica papaya papaya proteinase I (accession No. P00784), Lma; Leishmania major cathepsin L-like protease (accession No. AAB48120.1), Lme; Leishmania mexicana cysteine proteinase B precursor (accession No. P36400), Lp; Leishmania pifanoi cysteine proteinase 1 precursor (accession No. P35591), Tc; Trypanosoma cruzi cruzipain precursor (accession No. P25779), Tco; Trypanosoma congolense cysteine protease (accession No. AAA18215.1), Nf; Naegleria fowleri cysteine proteinase homolog (accession No. AAB01769.1), Pw; Paragonimus westermani pre-procathepsin L (accession No. AAB93494.1), Sm; Schistosoma mansoni cathepsin L precursor (accession No. Q26534), Sj; Schistosoma japonicum preprocathepsin cathepsin L (accession No. AAA87849.1), Pf; Plasmodium falciparum trophozoite cysteine proteinase precursor (accession No. P25805), Pv; Plasmodium vivax cysteine proteinase precursor (accession No. P42666), Fg; Fasciola gigantica cathepsin (accession No. AF112566_1), Fh; Fasciola hepatica secreted cathepsin L 1 (accession No. AAB41670.1).
Northern blot analysis
By Northern blot analysis of A. healyi poly (A+) mRNA, a single band of approximately 1.8 kb transcript was detected, thus supporting that the cloned cDNA should be a full-length (Fig. 5). To compare the level of mRNA expressions in pathogenic and nonpathogenic Acanthamoeba spp., total RNA from 9 different isolates were probed with the PCR product of AhCP1. As a control, the blot was reprobed with a fragment of rRNA cDNA of A. healyi, and intensities of the hybridization signals showed that comparable quantities of total RNA were present in all samples. There was no significant differences among isolates from infected cornea (KA/E1, KA/E2), contact lens storage cases (KA/L5) or infected brain (A. healyi OC-3A). Interestingly, two isolates from soil (KA/S8 and KA/CS) showed higher expression than other isolates (Fig. 5).
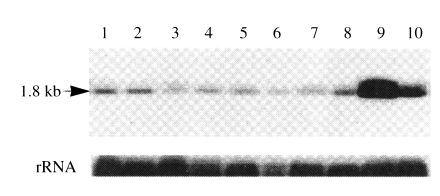
Northern blot analysis of AhCP1 transcript in different Acanthamoeba strains. Lane 1, Acanthamoeba sp. KA/E1; 2, Acanthamoeba sp. KA/E2; 3, Acanthamoeba sp. KA/L5; 4, A. castellanii Castellani; 5, A. healyi OC-3A; 6, A. polyphaga Jones; 7, A. castellanii Ma; 8, A. mauritaniensis 1652; 9, Acanthamoeba sp. KA/S8; 10, Acanthamoeba sp. KA/CS8.
DISCUSSION
In this study, we have isolated a cDNA (AhCP1) encoding a cysteine proteinase from A. healyi cDNA library. Analysis of the deduced amino acid sequence of AhCP1 from A. healyi suggested that it is translated as a preproenzyme. Karrer et al. (1993) classified C1 family cysteine proteinases into ERFNIN proteinases and cathepsin B-like ones based on the differences in the propeptide and in conserved amino acids of the mature proteins. We classified AhCP1 as ERFNIN proteinase because AhCP1 has the ERFNIN (EX3RX2(V/I)FX2NX3IX3N) motif which is highly conserved interspersed amino acids in propeptide region of proteinases in this class. In spite some ERFNIN proteinases of protozoa, for example, Trypanosoma brucei, Paramecium tetraurelia and Leishmania major, revealed substitution of amino acids residues in ERFNIN motif (Mottram et al., 1989; Volkel et al., 1996; Sakanari et al., 1997), AhCP1 showed perfect conservation of amino acid residues in the motif like mammalian cathepsin L. Several additional evidence of AhCP1 as ERFNIN proteinase were also found. In terms of amino acid sequence, many residues are known to be conserved within ERFNIN proteinases and different from those in cathepsin B-like proteinase. The seventh amino acid that is known to be an invariant tryptophan in mature ERFNIN proteinases was conserved in AhCP1. In contrast, in the case of cathepsin B-like proteinase, there is a consensus alanine at this position. Like other ERFNIN proteinases, the three amino acids that precede the active site of Gln19 were well conserved as VKN in AhCP1, whereas the cathepsin B-like proteinases are known to have IRD at these positions. The amino acid that precedes the active site Asn175 is a basic lysine (in most ERFNIN proteinase) or arginine (two cysteine proteinases from Tetrahymena thermophila) was also conserved as lysine in AhCP1. There is an invariant non-polar alanine at this position in the cathepsin B-like proteinases. Therefore this enzyme was regarded as a member of ERFNIN proteinases.
When the deduced amino acid sequence of mature enzyme was aligned to other known cysteine proteinase sequences, the sequence was strikingly homologous to mammalian cathepsin L or S (53-58% identity and 70-73% similarity) even though the highest homology was found in comparison with the sequence from Sarcophaga (identity 63% and similarity 73%). However, the enzyme shares 32-41% sequence identity and 47-57% similarity with other protozoan ERFNIN proteinases. Most interesting finding was that AhCP1 had very low sequence homology with AcCP2 of A. culbertsoni (Yun et al., 1999), and located in a different clade in the phylogenetic tree (Fig. 3). Ward et al. (1997) reported that phylogenetic analysis confirms the position of cysteine proteinase gene family of Giardia lamblia, one of the most primitive eukaryote as the first branch of the cathepsin B subgroup of peptidase family C1. Berti and Storer (1995) speculated that there was an explosive divergence and dispersion of cysteine proteinase genes coincident with the evolution of the first eukaryotic cells. These studies indicated that there had been two or more evolutionarily distinct groups of cathepsin L-like cysteine proteinases and that the evolutionary divergence of cathepsin L subgroups preceded the divergence of multicellular metazoa from unicellular protozoa. It can be also speculated that there have been two or more evolutionarily distinct ERFNIN proteinases subgroups in protozoa as in mammalian organisms. In addition to the structural similarity to mammalian cathepsin L, AhCP1 transcript size (1.8 kb) was the same as mammalian cathepsin L mRNA but much larger than those of several parasitic protozoan cysteine proteinase mRNA (1.0 kb). In the light of these characteristics, AhCP1 is an orthologous to mammalian cathepsin L.
Cysteine proteinases are now regarded to be biologically versatile as expected from evolutionary history of the molecules. The great majority of the cystein proteinases of C1 family are known to be lysosomal enzymes that function in intracellular protein degradation. However, some are released extracellularly and play histolytic roles in tumor metastasis or parasite invasion into host tissues. Although the exact functions of the Ahcp1 remains unclear yet, no significant difference of the expression between highly virulent and avirulent strains of Acanthamoeba indicated that Ahcp1 may be involved in intracellular protein degradation in this highly phagocytic predator cell. Different expression according to the origin of the amoeba or media (PYGC or PYC, data not shown)) also presented Ahcp1 as a digestive enzyme. Further study should be needed to determine the exact biological function of Ahcp1. We obtained the anti-Ahcp1 polyclonal antibody from a rabbit immunized with purified fusion protein expressed in E. coli. Ultrastructural study for the subcellular localization of AhCP1 can provide information on possible roles of the enzyme.
Notes
This study was supported by a grant (HMP-97-M-2-0016) of 1997, Good Health R&D Project, Ministry of Health and Welfare, Republic of Korea.
Note: Nucleotide sequence data reported in this paper is available in Genebank™ database under the accession number AF462309.