Abstract
We have studied the genetic differences among four isolates of Trichinella including a new strain of Trichinella spiralis (ISS 623) recently found from a human case who took a badger in Korea. Because they have a different host origin and came from geographically separated regions, we supposed the genetic pattern of the isolates might be different as had been previously reported. It was analysed by PCR-RFLP analysis of the rDNA repeat that can readily distinguish a species or strain from others. Isolated genomic DNA of each isolate of Trichinella larvae was amplified with ITS1 specific primers and digested with restriction endonucleases. The PCR product of ITS1 was confirmed using Southern blot analysis to be a 910 bp fragment. The restriction fragments of each isolate had variable patterns when it was digested with Rsa I only. According to the RFLP patterns, the estimated genetic divergence between each isolate was different. In conclusion, four isolates of Trichinella including a new strain of T. spiralis obtained from a Korean patient may have genetic differences in the ITS1 region and the Shanghai isolate was genetically more similar to the Japanese unknown isolate than others in the ITS1 region.
-
Key words: Trichinella, polymerase chain reaction, random amplified polymorphic DNA technique, ribosomal DNA
INTRODUCTION
Trichinosis continues to be a serious risk for man in some parts of the world. In Korea, human trichinosis outbreak was observed in 1997 at the first (
Sohn et al., 2000). The genus
Trichinella has a broad geographical background and host range, and different isolates can often display distinct biological characteristics. Variability within the genus
Trichinella has been reported in terms of morphology, infectivity, virulence, antigenicity, and enzyme polymorphism. During the last years, diversity has been analysed using parasitological, biological, morphological, immunological and biochemical (allozyme electrophoretic) parameters (
Flockhart et al., 1982;
Pozio, 1987;
Bolas-Fernandez and Wakelin, 1990).
DNA technology has also provided useful approaches to reveal genetic difference within the genus. Polymerase chain reaction (PCR)-based methods are popular technique recently (
Chambers et al., 1986;
Soule et al., 1993;
Minchella et al., 1994;
Zarlenga et al., 1999). It has been reported that PCR-RFLP (Polymerase chain reaction-restriction fragment length polymorphism) analysis of the rDNA repeat can readily distinguish a species or strain from others (
Boyd et al., 1989;
Zarlenga and Dame, 1992;
Gasser and Monti, 1997;
Nagano et al., 1999). The rDNA internal transcribed spacer (ITS) gene family consists of regions of highly conserved gene sequences that is flanked by transcribed and non-transcribed spacer regions, which may contain variations that can be useful for identifying the relatedness of strains. The ITS1 (internal transcribed spacer 1) PCR product used for a genetic marker to show differences among isolates studied here spans the ITS1 region of the rDNA repeat unit and includes most of the 5.8S gene and a portion of 18S gene (
Bowles et al., 1994). Here we have used the PCR-RFLP analysis of ITS1 rDNA and examined the patterns of four isolates of
Trichinella.
MATERIALS AND METHODS
Parasite
Worms of four isolates of Trichinella were maintained by numerous passage in laboratory SD (Sprague-Dawley) rats.; The Shanghai isolate from a domestic pig (presumed T. spiralis), the Thai isolate from a wild boar (T. spiralis, ISS 411), the Japanese unknown isolate from a raccoon dog (presumed T. spiralis) and Korean isolate recently found from a human who took a badger in Korea (T. spiralis, ISS 623). The Trichinella infected rats were sacrificed and digested in 1% pepsin-HCl at 37℃. After through several washing in distilled water, muscle larvae were collected under microscope avoiding contamination of host tissue debris and frozen at -70℃ until DNA isolation.
Isolation of genomic DNA
Genomic DNA from each isolate was purified as described by Rodriguez et al. (
1996) with minor modifications. Previously, the parasites were homogenized under liquid nitrogen with lysis buffer (50 mM Tris-HCl buffer pH 8.0, 50 mM ethylene diamine tetra-acetic acid (EDTA), 100 mM NaCl, containing 0.5% sodium dodecyl sulfate (SDS)). Then the homogenate was incubated with 100 µg/ml proteinase K (Sigma, USA) for 3 hours at 37℃. After an equal volume of phenol-chloroform extraction, the aqueous phase was digested with 150 µg/ml RNase (Sigma) for 30 minutes at 37℃. Additional phenol-chloroform extraction and chloroform extraction was performed subsequently. Total genomic DNA was precipitated by ethanol extraction and the DNA was resuspended in sterilized water. DNA concentration was determined by standard procedures.
A standard PCR reaction was performed using forward (5' GTCGTAACAAGGTTTCCGTA 3') and reverse (5' TCTAGATGCGTTCGAA(G/A)TGTCGATG 3') oligonucleotide. The internal transcribed spacer 1 (ITS1) specific primers were designed (
Bowles and McManus, 1993) by comparing conserved rRNA gene sequences from a range of organisms. Genomic DNA was amplified in a solution containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 5 mM MgCl
2, 0.2 mM of each dNTP and 2.5 units Taq polymerase (TaKaRa, Shiga, Japan). The following cycling conditions were used: 95℃, 30 sec (denaturation); 58℃, 1 min (annealing); 72℃, 1 min (extension) for 35 cycles on a thermocycler (MJ research). The PCR products were checked on agarose gel with ethidium bromide staining and transferred onto nylon membrane by standard capillary blotting method with 10 × SSC. Then the blotted membrane was hybridized with 5.8s rDNA probe made by PCR amplification with specific primers (forward: 5'-GGTACCGGTGGATCACT CGGCTCG-3', reverse: 5'-TCTAGATGCGTTCGAA(G/A)TGTCGATG-3'). It was detected by ECL™ detection system (Amersham, Buckinghamshire, England) as described in manual.
The 4-base recognizing restriction endonuclease for digestion of the ITS1 fragment were used.: Alu I, Cfo I, Msp I, Rsa I and Taq I. The PCR products were digested with several restriction endonucleases and analysed on agarose gel with ethidium bromide staining.
Analysis of ITS1 PCR-RFLP
Individual gel images of RFLP bands were analysed using phylogeny analysis program, RESTSITE v.1.2 (
Miller, 1994). The estimated genetic divergence was calculated using CALCD and the dendrogram was constructed by UPGMA v.2.0 (
Miller, 1994).
RESULT
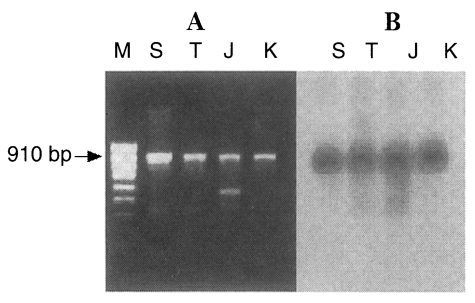
The PCR products of the ITS1 region of all
Trichinella isolates consisted of a fragment of approximately 910 bp and an additional band (525 bp) in the Japanese unknown isolate (
Fig. 1A). It was confirmed that the anticipated ITS1 product is indeed a 910 bp fragment by Southern blotting analysis with 5.8s rDNA probe that is included in ITS1 sequences (
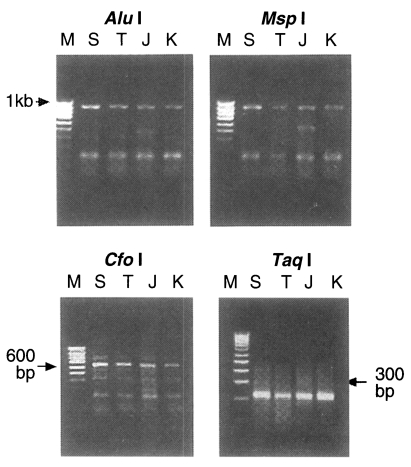
Fig. 1B). The PCR products were digested with
Alu I,
Cfo I,
Msp I,
Rsa I, and
Taq I. The RFLP pattern of four isolates digested using
Alu I and
Msp I were all the same at 910 bp uncut while digestion with
Cfo I and
Taq I produced the same two bands, 653 bp and 210 bp in every isolate respectively (
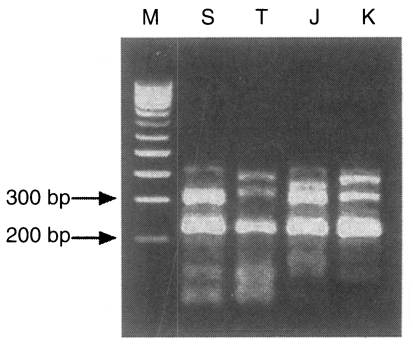
Fig. 2). But the pattern digested with one of 4 base recognizing enzymes,
Rsa I, distinctly showed the different patterns in ITS1 of four isolates. The Shanghai isolate showed a 308 bp band, the Thai isolate had two bands at 392 bp and 336 bp, the Japanese unknown isolate had 372 bp and 308 bp band, while the Korean isolate had bands of 385 bp and 313 bp (
Fig. 3).
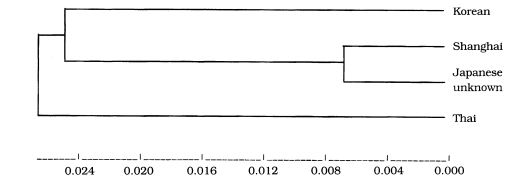
Estimated genetic convergence was analysed using RESTSITE v.1.2 (
Table 1). Genetic divergence between the Korean isolate and Shanghai isolate was 0.027218. It was revealed genetic divergence between the Korean isolate and the Japanese unknown isolate and between the Korean isolate and the Thai isolate was 0.03488 and between the Shanghai isolate and the Japanese unknown isolate was 0.008005. The phyllogenic dendrogram was drawn using UPGMA v.2.0 as shown in
Fig. 4.
DISCUSSION
In this study, we have shown the genetic difference of four isolates of
Trichinella using PCR-RFLP analysis. The genus
Trichinella has been tried to classify to species. Thus, nameless taxa (T5, T6 and T8) have been compared with already known species using various molecular and biochemical methods including RFLP (
Wu et al., 1999), RAPD (random amplified polymorphic DNA;
Bandi et al., 1993), SSCP (single-stranded conformational polymorphism;
Gasser and Monti, 1997) and isoenzyme assay, etc. (
La Rosa et al., 1982). These methods were also applied to the classification of many subspecies and isolates obtained from geographically distinct areas or different hosts. Dame et al. (
1987) grouped 18 isolates of the
T. spiralis from various origin to subspecies,
T. spiralis spiralis and
T. spiralis spp. by probe hybridization to restriction fragments of genomic DNA. If
T. spiralis isolates in the present study were compared with appropriate reference subspecies, they might have also been separated into the subspecies groups as known.
It is not surprising as heterogeneity among repeat units of rDNA is characteristic of eukaryotic DNA. Bowles et al. (
1994) genetically characterized several strains of
Echinococcus granulosus through PCR-RFLP analysis of the mitochondrial DNA as well as the nuclear ITS1 region and reported that the ITS1 region of rDNA repeat can distinguish the cervid strain from other strains of
E. granulosus. For the species
T. spiralis, it is apparent that the internal repeats of the IGS (intergenic spacer) and ITS region vary between isolates and are thus useful in distinguishing from others (
Boyd et al., 1989).
Here we tried to characterize the molecular genetic differences on the nuclear ITS1 region between the newly found Korean isolate and other isolates maintained experimentally. The results showed that the ITS1 PCR product amplified with ITS1 specific primers was identical in four isolates and Japanese unknown isolate had an additional band. Southern blot analysis confirmed that the band in all four isolates is the appropriate ITS1 product. And it's restriction enzyme fragments were different from each other when only one of the restriction enzymes, Rsa I, was reacted. It would appear that small genetic changes had occurred during the courses of evolution. However, it is not known at the present whether these differences are of any significance.
The estimated genetic divergence between the isolate obtained from a Korean patient and the Thai or the Japanese unknown isolate was comparatively higher than between the others. The result indicates that the Shanghai and Japanese unknown isolates are significantly genetically closer due to their proximity on the phyllogenic dendrogram. Thus the Shanghai isolate may be genetically more closer to the Japanese unknown isolate than others. It was thought that geographic location may contribute to the diversity in development of each isolate. Dame et al. (
1987) had determined that genetic differences do exist between sylvatic isolates and domestic isolates. In this vain, it is important to note that the origin of the Shanghai isolate is domestic (from the domestic pig) while the other isolates are known to be from wild hosts (the Korean strain originating from ingestion of a wild badger). However, there appears to be no significant relations with differing origins in our study. According to the dendrogram analysed, the Thai isolate may have evolved earlier than the others. This may be due to the geographic isolation of that isolate. In addition, it is suggested that Korean isolate is genetically distinct from the other three isolates presumed
T. spiralis (Shanghai, Thai and Japanese unknown isolate) from foreign countries and that three isolates also differ from one another.
In conclusion, it can be stated that the four isolates of the Trichinella genus display genetic difference in their respective ITS1 regions of rDNA. However the significance of these observations can only be determined when the rDNA of the various Trichinella isolates is sequenced. In turn, it will be possible to determine if the genetic differences found during RFLP analysis are due to differences in genetic development in the whole range of the gene or just due to point mutations. Further PCR-based molecular studies of other genes and sequences must be performed for proper molecular characterization and identification of subspecies of T. spiralis.
ACKNOWLEDGEMENTS
We would like to thank to Professor Chung Dong-Il, Department of Parasitology, Kyungpook National University for kindly providing the mice infected with Korean strain of T. spiralis.
References
- 1. Bandi C, La Rosa G, Comincini S, Damiani G, Pozio E. Random amplified polymorphic DNA technique for the identification of Trichinella species. Parasitology 1993;107:419-424.
- 2. Bolas-Fernandez F, Wakelin D. Infectivity, antigenicity and host responses to isolates of the genus Trichinella. Parasitology 1990;100:491-497.
- 3. Bowles J, McManus DP. Rapid discrimination of Echinococcus species and strains using a PCR-based method. Mol Biochem Parasitol 1993;57:231-239.
- 4. Bowles J, Blair D, McManus DP. Molecular genetic characterization of the cervid strain ('northern form') of Echinococcus granulosus. Parasitology 1994;109:215-221.
- 5. Boyd D, deVos T, Klassen G, Dick T. Characterization of the ribosomal DNA from Trichinella spiralis. Mol Biochem Parasitol 1989;35:67-72.
- 6. Chambers AE, Almond NM, Knight M, Simpson AJG, Parkhouse RME. Repetitive DNA as a tool for the identification and comparison of nematode variants: application of Trichinella isolates. Mol Biochem Parasitol 1986;21:113-120.
- 7. Dame JB, Murrell KD, Worley DE, Schad GA. Trichinella spiralis: Genetic evidence for synanthropic subspecies in sylvatic hosts. Exp Parasitol 1987;64:195-203.
- 8. Flockhart HA, Harrison SE, Dobinson AR, James ER. Enzyme polymorphism in Trichinella. Trans Roy Soc Trop Med Hyg 1982;76:541-545.
- 9. Gasser RB, Monti JR. Identification of parasitic nematodes by PCR-SSCP of ITS-2 rDNA. Molecular and Cellular Probes 1997;11:201-209.
- 10. La Rosa G, Pozio E, Rossi P, Murrel KD. Allozyme analysis of Trichinella isolates from various host species and geographical regions. J Parasitol 1982;78:641-646.
- 11. Miller JC. The RESTSITE v.1.2 Software package. 1994.
- 12. Minchella DJ, Eddings AR, Neel ST. Genetic, phenotypic and behavioral variation in North American sylvatic isolates of Trchinella. J Parasitol 1994;80:696-704.
- 13. Nagano I, Wu Z, Matsuo A, Pozio E, Takahashi Y. Identification of Trichinella isolates by polymerase chain reaction-restriction fragment length polymorphism of the mitochondrial cytochrome c-oxidase subunit I gene. Int J Parasitol 1999;29:1113-1120.
- 14. Pozio E. Isoenzymatic typing of 23 Trichinella isolates. Trop Med Parasitol 1987;38:111-116.
- 15. Rodriguez E, Nieto J, Castillo JA, Garate T. Characterization of Spanish Trichinella isolates by random amplified polymorphic DNA (RAPD). J Helminthol 1996;70:335-343.
- 16. Sohn WM, Kim HM, Chung DI, Yee ST. The first human case of Trichinella spiralis infection in Korea. Korean J Parasitol 2000;38:111-115.
- 17. Soule C, Guillou JP, Dupouy-Camet J, Vallet C, Pozio E. Differentiation of Trichinella isolates by polymerase chain reaction. Parasitol Res 1993;79:461-465.
- 18. Wu Z, Nagano I, Pozio E, Takahashi Y. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) for the identification of Trichinella isolates. Parasitology 1999;118(Pt2):211-218.
- 19. Zarlenga DS, Chute MB, Martin A, Kapel CM. A multiplex PCR for unequivocal differentiation of all encapsulated and non-encapsulated genotypes of Trichinella. Int J Parasitol 1999;29:1859-1867.
- 20. Zarlenga DS, Dame JB. The identification and characterization of a break within the large subunit of ribosomal RNA of Trichinella spiralis: comparison of gap sequences within the genus. Mol Biochem Parasitol 1992;51:281-290.
Fig. 1PCR products of the ITS1 region of four isolates of Trichinella (A). Southern blot analysis probed by 5.8S rDNA (B). M, 100 bp ladder; S, Shanghai isolate; T, Thai isolate; J, Japanese unknown isolate; K, Korean isolate

Fig. 2PCR-RFLP patterns of ITS1 region digested with Alu I, Msp I (uncut; upper pannel), Cfo I and Taq I (lower pannel). It shows a similar patterns among four isolates of Trichinella. M, 100 bp ladder; S, Shanghai isolate; T, Thai isolate; J, Japanese unknown isolate; K, Korean isolate

Fig. 3PCR-RFLP of four isolates digested with Rsa I. M, 100 bp ladder; S, Shanghai isolate; T, Thai isolate; J, Japanese unknown isolate; K, Korean isolate

Fig. 4Dendrogram of estimated genetic divergence. UPGMA v.2.0

Table 1.Estimated genetic convergence among four isolates of Trichinella using CALCD, RESTSITE v 1.2.
Table 1.
|
Korean |
Shanghai |
Japanese unknown |
Thai |
|
Korean |
|
|
|
|
|
Shanghai |
0.022328 |
|
|
|
|
Japanese unknown |
0.028797 |
0.006714 |
|
|
|
Thai |
0.028797 |
0.022328 |
0.028797 |
|