Genetic heterogeneity of Pneumocystis carinii from rats of several regions and strains
Article information
Abstract
Pneumocystis carinii is a major opportunistic pathogen which has been found in the lungs of a wide variety of mammalian host species, and the fact suggests the possibility of intraspecific variation. Until now, P. carinii from different mammalian species are differentiated as subspecies, and the rats are known to be infected by two subspecies. The present study investigated genetic heterogeneity of P. carinii isolates from two strains of rats in Korea and China by molecular karyotyping, RFLP and sequencing analysis. Karyotypes of P. carinii were grouped into three, two from two strains of rats in Korea and one from rats in China. However RFLP of PCR product of ribosomal and MSG gene of the P. carinii isolates showed same pattern. The sequence homology rates of α-tubulin DNA of the P. carinii isolates were 96% in Seoul Wistar rats, 93% in Seoul Sprague-Dawley rats, and 85% in Chinese Sprague-Dawley rats. The present finding confirmed that P. carinii from rats in Korea are grouped into two karyotype strains which are different from that of P. carinii from rats in China. The Chinese isolate shows a little different sequences of α-tubulin DNA.
INTRODUCTION
Pneumocystis carinii is a frequent opportunistic pathogen which causes pneumonia in patients with immunosuppressive disorders. It is a pathogen found mostly in the alveolar spaces, and the rate of infections has risen dramatically over the past years with the advent of aggressive immunosuppressive therapies and increasing infection with human immunodeficiency virus (HIV) (Wakefield et al., 1998).
The organism has appeared to infect a wide range of mammals. The hosts which had been reported to be infected by P. carinii were rats, mice, guinea pigs, hamsters, ferrets, rabbits, monkeys, dogs and cats (Walzer et al., 1989; Yoshida, 1993; Hong et al., 1999). The organisms from different host species are now classified as different subspecies (Stringer et al., 1997).
Genetic heterogeneity has been observed in P. carinii organisms isolated from different host species, suggesting that P. carinii infection is host-species specific. Analysis of the genetic heterogeneity of populations of P. carinii is contributing to better understanding of epidemiology and pathophysiology of this infection.
Heterogeneity of P. carinii has been reported in view point of not only antigenic variation but also genetic complexity (Hong et al., 1990; Stringer et al., 1993; Smulian et al., 1993). However, the study on the genetic heterogeneity of P. carinii is complicated by lack of in vitro culture system and occurrence of coinfections with several special forms or types in a single host (Vasquez et al., 1996).
In the present study, rat P. carinii isolates from different strains of rats and from several localities in Korea and one rat isolate from China were observed of their molecular karyotyping, restriction fragment length polymorphism (RFLP) and DNA sequencing to investigate the genetic diversity.
MATERIALS AND METHODS
Immunosuppression of rats for P. carinii infection
Rats were obtained from seven sources. Wistar strains of rats housed in Seoul 1 laboratory, and Sprague-Dawley (SD) rats housed in Seoul 1 and Seoul 2 laboratories, SD rats from Chunchon, Chungju, Kwangju, and Shanghai, China. Each group of rats immunosuppressed by weekly injection of 10 mg/kg methylprednisolone (Depomedrol®, Upjohn Korea Ltd.). Drinking water was supplemented with 1 mg/ml of tetracycline. After 8 weeks, all of the rats were sacrificed and their lungs were removed and smeared for examination of infection intensity. Heavily infected lungs were stored in freezing media at -70℃ until preparation for pulse-field gel electrophoresis and DNA isolation.
Preparation of P. carinii organisms for pulsed-field gel electrophoresis (PFGE)
The lungs were homogenized and prepared for pulsed-field gel electrophoresis as previously described. After homogenization in a laboratory blender (Stomacher; Tekmar Inc., Cincinnati, Ohio, USA), the preparations were filtered through gauze to remove large particulate host material, treated with 0.85% ammonium chloride, and then passed at least twice through 10-µm-pore-size filters (Mitex, Millipore Corp., Bedford, Mass.). Each preparation of P. carinii was kept separate. The P. carinii preparations were monitored for other microbes by using microscopic methods. P. carinii organisms were treated with 10 µg/ml DNase I (Boehringer Mannheim, Indianapolis, Ind., USA) at for 30 min 37℃ to digest extracellular DNA. The organisms were washed successively with 0.25 and 0.125 M EDTA and collected by centrifugation. Then organisms were embedded by reconstituting organism pellets in 0.125 M EDTA in 0.8% low-meltingpoint agarose (Sigma Chemical Co., St. Louis, Mo., USA).
Gel-embedded cells were treated with 0.25 mg/ml proteinase K (Boehringer Mannheim Biochemicals, Indianapolis, Ind., USA) and 1% sarkosyl at 55℃ for up to 12 hours. Digested agarose plugs were stored in 0.5 M EDTA (pH 9.0) at 4℃.
Molecular karyotyping by CHEF (contour clamped homogeneous electric field gel electrophoresis)
Pulse field gel electrophoresis was performed using contour clamped homogeneous electric field (CHEF) analysis (CHEF DR II, BioRad, USA). Embedded organisms were electrophoresed through 1.3% agarose gels (FMC SeaKem agarose (FMC Bioproducts, Rockland, Maine, USA) at 14℃ for 3-40 hours at 6V/cm with gradual switching from 150 to 60 sec.
Restriction fragment length polymorphism (RFLP) and DNA sequencing analysis
P. carinii DNA samples were obtained from the same agarose blocks used for karyotyping. PC S3 (5'-ACTGATCCTTCCCTCCTGPC-3') sense primer of ribosomal DNA (rDNA), and PC AS4 (5'-AATAACCCA TCACCAG-3') primer for PCR amplification, corresponding to 1019 bp of rat P. carinii ribosomal DNA, and PC S8 (5'-GAGTGA TTGTTATGGGG-3') and PC AS8 (5'-GCCAAAAGGTGTTTCTC-3") to 600 bp of rat P. carinii MSG gene. The samples ware amplified in a DNA thermal cycler 9600 (Perkin Elmer, USA) for 30 cycles (94℃, 30 sec, 55℃, 30 sec, 72℃ 30 sec). Each reaction was electrophoresed for identification of products.
PCR products were reacted by 17 different restriction enzymes (Boehringer-Mannheim Biochemicals, Indianapolis, Ind., USA), and performed sequencing the products of PC S5 and PC AS5 α-tubulin DNA using an automated sequencer model 377 (Perkin Elmer, USA). The nucleotide sequence data were analysed for sequence homology.
RESULTS
Karyotyping of P. carinii by strains of rats by CHEF
Three karyotype patterns of P. carinii were found from all groups of rats (Fig. 1, Fig. 2). Pattern of P. carinii from Wistar strains of rats was different with that of SD strains of rats. P. carinii from SD strains in China had another different karyotype pattern. Between P. carinii from SD strains from various regions in Korea, the karyotype showed similar patterns.
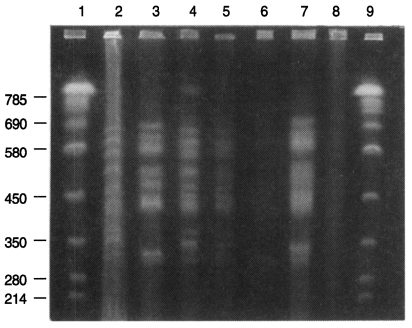
A CHEF gel of 1.3% agarose in 0.5X TBE, ethidium bromide stained. Running parameters are initial A time is 15 seconds and final A time is 60 seconds, A/B ratio is 1, 6 V/cm for 40 hours. lane 1. Saccharomyces cervisease AB 975; lane 2. W rats' P. carinii from Seoul; lane 3. SD rats' P. carinii from China; lane 4. SD rats' P. carinii from Seoul 1; lane 5. SD rats' P. carinii from Seoul 2; lane 6. SD rats' P. carinii from Chunchon; lane 7. SD rats' P. carinii from Chungju; lane 8. SD rats' P. carinii from Kwangju; lane 9. Saccharomyces cervisease AB 975.
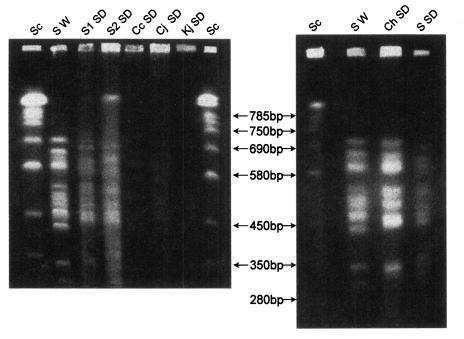
A CHEF gel of 1.3% agarose in 0.5X TBE, ethidium bromide stained. Running parameters are initial A time is 15 seconds and final A time is 60 seconds, A/B ratio is 1, 6 V/cm for 40 hours. SC. Saccharomyces cervisease; S W. W rats' P. carinii from Seoul; Ch SD. SD rats' P. carinii from China; S1 SD. SD rats' P. carinii from Seoul 1; S2 SD. SD rats' P. carinii from Seoul 2; Cc SD. SD rats' P. carinii from Chunchon; Cj SD. SD rats' P. carinii from Chungju; Kj SD. SD rats' P. carinii from Kwangju.
Restriction analysis of PCR amplification products of rat P. carinii DNA
Ribosomal primer (S3/AS4) : All of the isolates produced the amplification products of 1019 bp. It was obtained 1019 bp. After treated all with 14 restriction enzymes, all the same restriction patterns showed between the isolates (Table 1, Table 2).
MSG primer (S8/AS8) : It was obtained 600 bp of expected amplification products. After treated all with 17 restriction enzymes, all the same restriction patterns showed between the isolates (Table 1, Table 2).
Sequence analysis of PCR products of P. carinii α-tubulin DNA
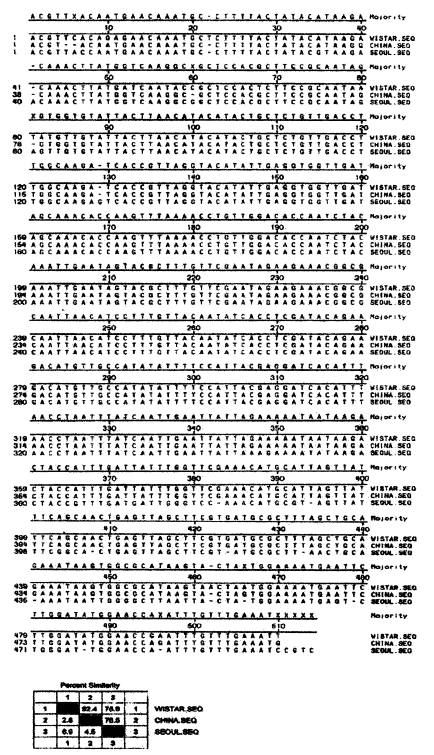
All of the 3 isolates showed same sized PCR products of 512 bp of P. carinii α-tubulin DNA. The products were completely sequenced by ABI prism automated DNA sequencer (Perkin-Elmer, USA). The sequences were compared with P. carinii α-tubulin DNA from genebank database (Fig. 3). The homology percentages of P carinii is 96% in Seoul Wistar, 93% in Seoul, 85% in China.
DISCUSSION
The chromosomal bands of rat Pneumocystis carinii resolved up to 15 from 290 to 710 kb. The karyotype pattern and genomic size of 1×107 bp are same as previously estimated although some isolates were very faintly resolved (Hong et al., 1999).
However, total 6 present isolates of rat Pneumocystis in Korea were grouped into two karyotype patterns. One pattern was of Pneumocystis from Wistar (W) rats in Seoul and the other was of P. carinii from Sprague-Dawley (SD) rats in Seoul. The pattern from SD rats showed one distinctive band of 710 kb size which was absent in the isolate from W rats. The SD rats in Seoul were supplied by two independent vendors but their P. carinii karyotype was same. One vendor also supplied W rats as well as SD rats but the two strains harbour P. carinii of different karyotypes.
P. carinii isolates from all of SD rats which were locally supplied and reared in Chunchon, Chungju and Kwangju showed the same pattern from SD rats in Seoul. Therefore the karyotype of rat P. carinii in Korea may be grouped into two by the strain of rats, W and SD.
The P. carinii isolates from SD rats which were experimentally infected in China showed a little different karyotype pattern. The most significant difference was missing one chromosome band of 380 kb in the Chinese isolate.
The chromosome molecules of P. carinii from rats in USA range from 290 to 710 kb (Hong et al., 1990; Cushion et al., 1993). The chromosomes of P. carinii in the present study showed similar range of size and number with those of USA isolates. There were 3 to 5 large chromosomes from 570 to 710 kb, 6 middle ones from 420 to 520 kb, and 2 to 4 small ones from 290 to 370 kb.
Basically P. carinii is cosmopolitan over the world. However, there are several karyotype strains in the world. The P. carinii from different host species shows different karyotype pattern (Hong et al., 1999), but the rats are known to be co-infected by P. carinii of different patterns (Hong et al., 1990; Cushion et al., 1993).
The present study revealed two karyotype strains of P. carinii from rats in Korea and one strain from rats in China. The karyotype pattern from SD rats in Seoul is very similar with that of P. carinii carinii (Cushion et al., 1997) but it is hard to match the other two patterns with that of the other subspecies, P. carinii rattus. The two present strains are still possibly either P. carinii carinii or P. carinii rattus, and this should be a subject of further studies.
The two karyotype strains of rat P. carinii were found to be derived from strains of rats not from geographical localities. Formerly the karyotype was regarded as depending upon the rat colony rather than the strain. However the W and SD rats which were supplied from the same animal vendor showed different P. carinii karyotypes while SD rats from different animal colonies showed the same karyotype pattern. The finding strongly suggests that P. carinii may be transmitted through the generation of rats in the closed colony from the primary ancestor of the colony. All of the rat vendors in Korea and China have introduced their ancestor rats from USA or Japan. The SD rats from several localities in Korea may have shared their ancestor from one colony in USA but the Chinese SD rats may not.
Data supporting biological meaning of karyotype strains of P. carinii are still insufficient. Though P. carinii from rats in Korea and China are grouped into 3 karyotype strains, the present data on RFLP of the PCR products of 16S ribosomal and MSG gene of the 3 strains showed no strain differences. The PCR product of ribosomal gene was 1000 bp and that of MSG gene was 510 bp. The sequence of 500 to 1000 bp of two genes looks still short to identify definite strain differences of P. carinii. More genes and longer sequences are to be compared for definite biological identification of the karyotype strains.
P. carinii from different mammals are known to express different antigenicity (Bauer et al., 1993). Isolates of P. carinii from humans are also antigenically complicated (Sinclair et al., 1991). Of course karyotype strain is still possibly indifferent from the antigenicity. Also whether other genetic differences may be correlated with the karyotype strain is to be determined.
The karyotype difference may be induced by simple translocation of a telomere of a chromosome to another one or by duplication of a telomere (Lundgren at al., 1990). In that situation, the coding sequences of expression may be unchanged but the chromosomal size may be changed. However, the karyotype of a rat colony was found rather stable for several years (Hong et al., 1990; 1992b; 1997). This stability may be used to trace any specific pedigree of P. carinii or also the host (Hong et al., 1992b). It may be an epidemiological tracer.
The sequence homology rate of α-tubulin gene DNA of rat P. carinii was 93% in SD rats and 96% in W rats, which may be insignificant. Contrary to this, the P. carinii from Chinese SD rats showed 85% homology rate which should be regarded as low homology because α-tubulin DNA is a gene of active translation (Wada et al., 1993). It is necessary to compare the antigenicity of the Chinese isolate.
In conclusion, there were two karyotype strains of rat P. carinii in Korea. The Pneumocystis strain was correlated with the strains of host rats. One Chinese isolate also showed a different karyotype and low homology rate of α-tubulin DNA sequences. The karyotype may be a tracer of any specific strain or isolate of Pneumocystis.
Notes
The present study was supported by the grant for Basic Medicince Research Fund from Ministry of Education, Republic of Korea (1995-1997).