PCR Detection and Molecular Characterization of Pentatrichomonas hominis from Feces of Dogs with Diarrhea in the Republic of Korea
Article information
Abstract
Pentatrichomonas hominis is considered a commensal protozoan in the large intestine of a number of mammalian hosts, such as cats, dogs, and non-human primates. The resulting infections, which can induce diarrhea, have been attributed to opportunistic overgrowth of P. hominis. This study was performed to confirm the P. hominis infection and its molecular characterization from the feces of puppies with diarrhea. Fecal samples were obtained from 14 German shepherd puppies with diarrhea over 1 week (7 females and 7 males, 2-9 months of age) residing on a dog farm in August 2007. Species-specific PCR assay identified P. hominis 18S rRNA genes in 3 of the 14 puppies (1 female and 2 males; 1 aged 2 months and 2 aged 9 months). This phylogenetic analysis established that P. hominis belonged to the 1st clade, which is comprised of Bos taurus and Felines.
INTRODUCTION
Trichomonads, such as Pentatrichomonas hominis, are obligate protozoan symbionts found in vertebrates and are considered to be among the most primitive of eukaryotic organisms. They are characterized morphologically by multiple anterior flagella and a single recurrent flagellum that functions as an undulating membrane [1]. P. hominis inhabits the large intestine of a number of mammalian hosts, including cats, dogs, non-human primates, and pigs [2-6]. Opportunistic overgrowth of P. hominis can cause disease, typically involving diarrhea [7,8]. The potential zoonotic risk of this organism remains to be determined, in part due to the limited number of reports. Recently, sensitive and highly specific oligonucleotide probes enabling the identification of P. hominis were reported [9], as were the optimum reaction conditions and detection limits of polymerase chain reaction (PCR)-based identification of P. hominis in DNA extracted from canine feces [10]. The present report is the first identification of P. hominis infection in puppies with diarrhea in the Republic of Korea. As well, the taxonomic status of the P. hominis isolates are compared with those isolated elsewhere.
MATERIALS AND METHODS
Sampling
The (fecal) samples were obtained (from the dogs with diarrhea) from 14 puppies over a one-week period in August 2007, residing on a dog farm located in Yangju-si, Northern Gyeonggi province, Republic of Korea. All were German shepherd puppies (7 males, 7 females) ranging in age from 2-9 months, and all had received the regular general vaccinations. All stool samples were examined by light microscopy in veterinary clinic, and frozen and stored at -20℃ prior to DNA extraction. And then all samples were moved to our laboratory within 1 week and DNAs of them were extracted.
DNA extraction and PCR analysis
Genomic DNA was extracted from 200 mg of each fecal sample using a QIAamp DNA Stool Mini Kit (Qiagen, Valencia, California, USA) using the manufacturer's instructions. P. hominis-specific primers used were Th3 (5'- TGT AAA CGA TGC CGA CAG AG -3') forward and Th5 (5'-CAA CAC TGA AGC CAA TGC GAG C-3') reverse [10]. PCR involved 50 cycles of 1 min of denaturation at 95℃, 1 min of annealing at 64℃, and 2 min of extension at 72℃ [10]. The first cycle was preceded by 5 min of denaturation at 95℃ and the last cycle was followed by 5 min of final extension at 72℃. The volume was adjusted to 20 µl with Milli-Q ultrapure water (Millipore, Billerica, Massachusetts, USA). One microliter of DNA extract was added to each PCR premix kit (Bioneer, Seoul, Korea). Upon completion of PCR, 10 µl of each amplified specimen was analyzed by electrophoresis in a 2% agarose gel in Tris-acetate-EDTA buffer (pH 8.5). The gel was stained with ethidium bromide (0.5 µg ml-1; Sigma-Aldrich, St. Louis, Missouri, USA) and was photographed under short wavelength ultraviolet light. The PCR products were purified by QIAquick PCR purification kit (Qiagen) and sequenced by a commercial laboratory (Macrogen, Seoul, Korea).
Sequence alignment and phylogenetic analysis
The partially sequenced P. hominis 18S ribosomal RNA gene was accessed from the GenBank data library of the National Center for Biotechnology Information. Alignment of nucleotide sequences were performed using the Clustal X 1.81 program [11]. Phylogenetic trees for Pentatrichomonas spp. were carried out by the neighbor-joining method using MEGA 3.1 [12]. Local bootstrap probability was calculated from 1,000 replications.
RESULTS
Microscopic examination and PCR analysis
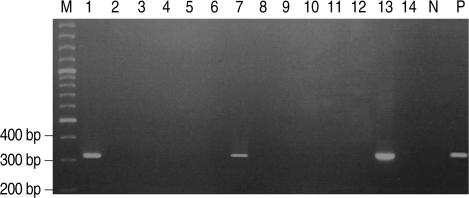
Stool wet mounts of 14 puppies with diarrhea reared in a dog farm were observed by light microscopic examination. Trichomonad-like organisms were evident in samples from three of the 14 puppies (21.4%). One of the three puppies was female and the other two were males. One puppy was 2-months-old and the other two were 9-months-old (Table 1). All three visually positive samples yielded a 339-bp amplified P. hominis specific DNA (Fig. 1).

The results of microscopy and PCR for Pentatrichomonas hominis infection in 14 German shepherd puppies with diarrhea over 1 week in August 2007
Phylogenetic analysis
The partially sequenced 18S rRNA gene of P. hominis amplified by PCR was aligned and the sequence compared to the nucleotide sequences of P. hominis (GenBank accession number: AF124609), P. hominis (ac: DQ412643), P. hominis (ac: DQ412642), P. hominis (ac: DQ412641), P. hominis (ac: DQ412640) and P. hominis (ac: DQ899948). These sequences were almost identical, differing only in alignment at 1-7, 9, 17, 29, 144, 292 and 302 (Fig. 2). Most notably, query coverage was 96% compared with P. hominis 2003-5007 (DQ412643) in the GenBank NCBI database using BLAST X. The phylogenetic analysis identified three well-resolved terminal Clades supported by high bootstrap values (Fig. 3). P. hominis identified in this study was assigned to the 1st Clade, where the host of origin comprises animals such as Bos taurus and felines [10,13].
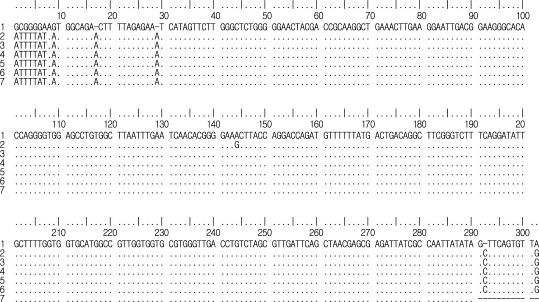
Comparison of alignment in 18S rRNA gene sequences of P. hominis amplified in the present study with others. (1) P. hominis in the present study; (2) P. hominis (GenBank accession number: AF124609); (3) P. hominis (ac: DQ412643); (4) P. hominis (ac: DQ412642); (5) P. hominis (ac: DQ412641); (6) P. hominis (ac: DQ412640); (7) P. hominis (ac: DQ899948). Dots indicate nucleotides identical to those of P. hominis and dashes are gaps inserted to optimize alignment. Numbering is based on the data registered in GenBank.

Neighbor-Joining (NJ) tree of Pentatrichomonas hominis from 18S rRNA gene sequences. P. hominis analyzed in this study are marked in rectangle. Data for tree construction obtained from GenBank Data Libraries of the NCBI. The length of each branch is proportional to the amount of evolutionary distance between the different species. Scale bar indicates 0.2 substitutions (corrected) per base pair.
DISCUSSION
P. hominis is a trichomonad species found in humans and animals. Although P. hominis inhabits the large intestine of mammalian hosts and is considered to be a commensal [14], their molecular characterization and pathogenicity are ill-understood. The organism is characterized morphologically by five anterior flagella and a single recurrent flagellum that functions as an undulating membrane [15,16]. In this study, trichomonad-like parasites were observed in three of 14 fecal samples by light microscopy. A more detailed morphological examination, which is possible using a recently reported methylene blue staining regimen, was not undertaken because the fecal samples were frozen prior to refer to our laboratory.
We used the primer pair Th3 and Th5, which have 100% sequence identity with a canine isolate of P. hominis 18S rRNA genes [10]. P. hominis infection rate in the present study, based on the aforementioned light microscopy examination, was 21.4% (3/14; one female and two males). As previously reported, dogs presenting with trichomoniasis and diarrhea range widely in age from < 13 weeks to 6 months [10,17-20]. In this case, the affected puppies were 2-months-old (n = 1) and 9-months-old (n = 2), which is similar to previous studies in age. Interestingly, the parents of the affected puppies and other adult dogs in the same dog farm displayed no symptoms of diarrhea and were PCR-negative for P. hominis (data not shown). This is similar to a previous report [21]. It may be that puppies are more susceptible to P. hominis infection than adults, although this remains to be rigorously ascertained.
Trophozoites of P. hominis reproduce by binary fission and undergo direct host-to-host transmission without formation of environmentally stable cysts [1]. Because all of the 14 puppies we sampled experienced diarrhea at a similar time, the spread of P. hominis infection in the dog farm presumably occurred by contacting of the puppies during their regular communal exercise periods. The affected puppies were successfully treated empirically with metronidazole. In spite of the general vaccine program, whether P. hominis infection was a cause of diarrhea over one week in this case could be concluded from these observations alone. More detailed epidemiological studies of P. hominis infections should be required.
According to the phylogenetic tree, P. hominis (ac: AF124609), P. hominis (ac: DQ412643), P. hominis (ac: DQ412642), P. hominis (ac: DQ412641), P. hominis (ac: DQ412640) and P. hominis (ac: DQ899948) formed the 1st Clade. P. hominis (ac: AY349178), P. hominis (ac: AY245137), P. hominis (ac: AF342741), P. hominis (ac: AF156964), P. hominis (ac: AY758392) and P. hominis (ac: U86616) formed the 2nd Clade. P. hominis (ac: AF288745), P. hominis (ac: Z18250), P. hominis (ac: Z18259), P. hominis (ac: AY886880) and P. hominis (ac: AY886879) formed the 3rd Clade. P. hominis (ac: Z18258) was placed independently between the three Clades. P. hominis (AY245137) and P. hominis (AY886880 and AY886879), which are normal human commensals, were assigned to both the 2nd and 3rd Clades, but the P. hominis identified in this study was assigned to the 1st Clade, where the host of origin comprises animals such as Bos taurus and felines [10,13]. These natural hosts are consistent with the presence of P. hominis found in puppies with diarrhea in the present study; the dog farm is located in a mountainous and rural setting where contact with wild animals naturally harboring the bacterium could easily occur. Although the pathogenic potential of P. hominis in dogs remains unknown, the present study is the first to establish a molecular identification of P. hominis in puppies with diarrhea and confirm the taxonomic status of P. hominis in the Republic of Korea. Further studies concerning morphological identification, clinical isolation and confirmation of pathogenicity of P. hominis are required.
ACKNOWLEDGEMENTS
This work was supported by a grant (NIH-4847-302-210-13, 2008) from the National Institute of Health, Korea Centers for Disease Control and Prevention.