Two new genotypes of Plasmodium vivax circumsporozoite protein found in the Republic of Korea
Article information
Abstract
The gene encoding Plasmodium vivax circumsporozoite protein (PvCSP) exhibits polymorphism in many geographical isolates. The present study was designed to investigate polymorphism in PvCSP gene of P. vivax isolates in Korea. Thirty isolates, obtained from indigenous cases in Yonchon-gun, Kyonggi-do in 1997, were subjected for sequencing and RFLP analysis of the repeat and post-repeat regions of PvCSP gene and two genotypes (SK-A and SK-B) were identified. The genotype of 19 isolates was SK-A and that of 11 isolates was SK-B. Although the number of 12-base repeats present in SK-A was three while two were found in a Chinese strain CH-5, the repeat sequence of SK-A was identical to that of CH-5 except for one base substitution. Compared with known data there was no identical isolates with SK-B, but the sequence of SK-B was similar to that of a North Korean (NK) isolate. These results indicate that two genotypes of PvCSP coexist in the present epidemic area of Korea and the present parasite may originate from East Asia. RFLP would be useful to classify genotypes of P. vivax population instead of gene sequencing.
INTRODUCTION
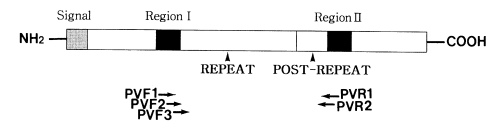
The circumsporozoite protein (CSP), a stage specific immunodominant surface antigen, is expressed during the prehepatic sporozoite stage of all malaria species (Di Giovanni et al., 1990; Escalante et al., 1995). The CSP gene sequence has been characterized in human malaria parasites, Plasmodium falciparum, P. vivax, and P. malariae (Dame et al., 1984; Arnot et al., 1985; Qari et al., 1994). These studies have shown that the length and the sequence of repeat regions differ considerably among malaria species. In addition, comparative studies on the intraspecific variation in the CSP gene sequence have established the wide genetic diversity in P. falciparum and P. vivax parasite populations (Lackyer et al., 1987; Qari et al., 1992; Jongwutiwes et al., 1994; Rongnoparut et al., 1995). The diversity was based on the repeat regions of the P. vivax CSP (PvCSP). The CSP contains about 20 nonapeptide repeats in repeat regions of P. vivax. So far, two families of this gene have been classified based on the consensus sequence of the following: Type I and Type II repeats represented in amino acid sequences which are GDRADGQPA and ANGAGNQPG, respectively (Rogenberg et al., 1989; Qari et al., 1991).
Mann et al. (1994) classified the isolates which have type I repeats into several groups based on the repeat structure. In his work, it was observed that, although there was a potential for many combinations of repeats, a limited number occurred and the combinations were relatively stable in both locality and the time. Thus, he postulated that the classification based not only on the repeat structure, but it could be extended their geographical origins.
Meanwhile, P. vivax malaria has reemerged and has been regarded as one of the major public health problems in Korea (Lee et al., 1998; Chai, 1999; Kho et al., 1999). However, the exact reason for the reemergence is still unclear and a few gene structures of the parasites are available in the present epidemic. In this paper, we present DNA sequence data on the PvCSP gene of the isolates from the Republic of Korea. To trace the geographical origin of the parasites, we compared the obtained data to the previous published sequencing data.
MATERIALS AND METHODS
Preparation of parasite DNA
Thirty blood samples were collected from P. vivax patients who were detected in Yonchon-gun, Kyonggi-do in 1997. These patients were diagnosed by microscopic examination of their peripheral blood smears.
The parasite DNA was extracted from the blood according to a modified method described by Arai et al. (1994). Briefly, blood cells were incubated in 0.51% saponin solution at 37℃ for 20 min, and then the pellet was washed in TSE solution [10 mM Tris (pH 8.0), 1 mM EDTA, 10 mM NaCl]. After digesting with proteinase K, phenol: chloroform: isoamyl alcohol (25:24:1) were added. After centrifugation at 6,000 g for 10 min the DNA was precipitated with absolute alcohol. All DNA samples were stored at -70℃ until use.
Polymerase chain reaction (PCR)
To amplify the CSP gene, two sets of forward and reverse primers were synthesized (Fig. 1). The first set of primers were 21- and 21-mers: 5'-CCAGATGACGAGGAAGGAGAT-3' (PVF1) and 5'-CGCCGAAAATATGGATGTCA-3' (PVR1). The second set of PVF2 and PVR2 primers were nested to the first set. These were 19- and 24-mers: 5'-AAATAAGCTGAAACA ACCA-3' (PVF2) and 5'-CACTCCACAGGTTACACTGCATGG-3' (PVR2). These primers amplified the domains, including the repeat and post repeat variable regions of CSP. Two µl of extracted DNA solution, 10 pmol of one set of primers, 0.5 U Taq polymerase (Perkin Elmer, USA) and 0.2 mM dNTPs were added to a reaction volume of 20 µl. The cycling conditions were as follows: first denaturation at 95℃ for 5 min; 40 cycles of 94℃ for 1 min, 55℃ for 1 min and 72℃ for 1 min; final extension at 72℃ for 10 min. A sample of 2 µl of the first PCR product was transferred to a new tube, followed by the addition of the primers, fresh Taq polymerase and fresh dNTPs. The nested PCR reaction was performed as described above.
Sequencing and analysis
The PCR products were electrophoresed on a 1.2% agarose gel. The amplified DNA was purified using a QIAEX II gel extraction kit (Qiagen, USA) following the manufacturer's instruction. The nucleotide sequence was determined by dideoxynucleotide chain termination method using Sequenase kit (ABI PRISM Dye Terminator Cycle Sequencing Core Kit, Perkin Elmer) and an automated DNA sequencer (Applied Biosystems model 377A, Perkin Elmer). The PVF3 (5'-AAAAAGGATGGAAAGAAAG-3') and PVR2 primers were used for direct sequencing. The manual sequencing was processed using T7 Sequenase version 2.0 kit (Amersham, UK) with PVR2 primer.
The DNA sequencing data were analyzed using DNASIS (Hitachi, Ver. 2.5, Japan) and the BLAST programs of the NCBI databases (Bethesda, USA). Also our sequence data were further compared with published sequences of P. vivax isolates (Mann et al., 1994).
RFLP analysis
The amplified CSP fragments were digested using PvuII restriction enzyme (Promega, USA) at 37℃ for 2 hr and the digested fragments were separated by polyacryamide gel electrophoresis.
RESULTS
Amplification of the PvCSP gene fragment
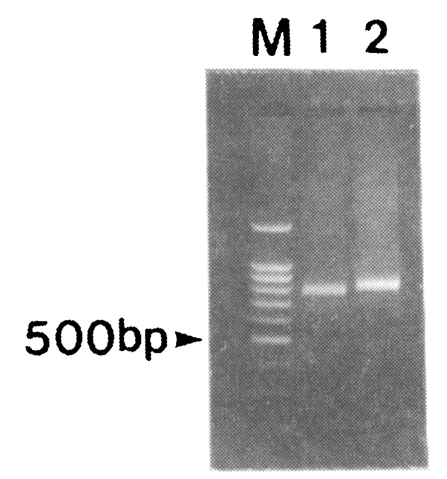
Amplification of the fragment of CSP was attempted for 30 blood samples obtained from patients in the Republic of Korea. After the two rounds of PCR, 791 and 833 bp bands were amplified (Fig. 2). Nineteen out of thirty isolates produced 791 bp band while eleven isolates produced 833 bp band. No bands were amplified from non-infected donors (data not shown).
Sequence analysis
The repeat and post-repeat regions of the PvCSP gene were sequenced and the two genotypes (SK-A and SK-B) included type I repeats, GDRADGQPA. The number of 27 base repeats was 18 for SK-A and 20 for SK-B (Fig. 3). A 36-base insert was observed immediately at the 3' to the repeat in both genotypes. The number of the 12-base repeat following the insert was two for SK-A and three for SK-B (Fig. 4). In the repeat region, SK-A was identical to that of CH-5 with the exception of one base substitution (Mann et al., 1994). However, the number of 12-base repeats was three for SK-A and two for CH-5. In comparison to the published data, there was no identical isolate of SK-B. However, the sequence of SK-B was similar to that of a known North Korean (NK) isolate (Mann et al., 1994).

The nucleotide sequences of the PvCSP gene of South Korean isolates compared to the NK and China groups. Repeats are numbered and the sequences read from 5' to 3', starting at the first base of the first numbered tandem repeat to the last codon in the last numbered repeat. The predicted amino acid is given above the nucleotide sequence. The sequences are compared to the NK isolate and CH-5 (Mann et al., 1994).

Variation in the nucleotide sequences of end of the last 27-bp repeat, 36-bp insert and 12-bp repeat. The sequence of post-repeat region of PvCSP was compared with NK, CH2-7, PH79, VK210, SOL-83, SOL-101, Belem and NYUThai (Mann et al., 1994). The bold numbers indicate 12 bp repeat. Shaded squares reveal identical sites.
RFLP analysis
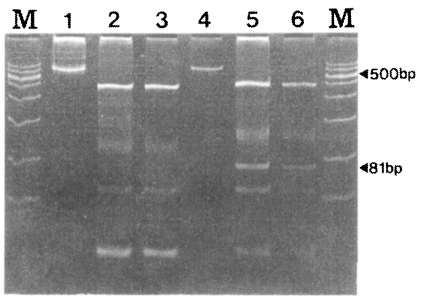
To distinguish between SK-A and SK-B, RFLP using PvuII was performed with 30 isolates. As in Fig. 5, SK-A showed fragments with sizes of 436, 100, 54, and 24 bp while SK-B showed fragment of 451, 127, 81, 54 and 27 bp. The 81 bp fragment was shown only by SK12 (Fig. 5).
DISCUSSION
The genotype of the present 30 South Korean isolates, which was based on the sequence data of the polymorphic regions between the repeat and post-repeat regions of the PvCSP gene, showed that two genotypes, SK-A and SK-B, coexist in the present epidemic area of Korea. The coexistence of several genotypes in the same endemic area was already observed in China. Six different genotypes were observed in the Hongmao coal mine near the border of Gaungxi and Guizhou provinces (Mann et al., 1994). In conjunction with this finding, it is possible that several other PvCSP genotypes exist in Korea; therefore, a sequence analysis of the PvCSP gene of more isolates may be a topic of further study.
The isolates of P. vivax have been classified by DNA sequences of the repeat and post-repeat regions (Mann et al., 1994). According to the Mann's criteria, although the sequences were not identical to those of the other known isolates around the world, the sequences of the present South Korean isolates were similar to those of CH-5 (originated from China) and the NK isolates which are classified as the East Asian group. Therefore, the present data strongly suggested that the recent reemergence of P. vivax in South Korea could have originated from East Asia.
It is believed that the variation of PvCSP occurs due to the genetic recombination between the variants (Arnot et al., 1988). Since P. vivax malaria had been endemic for many centuries, many variants should have been produced in the Korean peninsula. The isolates observed in the present study could be introduced from abroad. However, the possibility that they could be produced by the process of genetic recombination within the Korean peninsula long before the present epidemic cannot be excluded. The NK strain is so far the only isolate from North Korea which was isolated in 1953 and maintained at a Moscow psycho-neurological hospital in patients undergoing malaria therapy (Collins et al., 1985). In 1968, the NK strain of P. vivax was transported to a laboratory in London where the strain had been maintained using anopheline mosquitoes and splenectomized chimpanzees for about 30 years (Collins et al., 1985). The NK strain was isolated from infected Saimiri sciures monkey and its PvCSP gene was sequenced (Arnot et al., 1988). However, the possibility of the alteration of its DNA structure due to the host change or the long passage in the laboratory cannot be excluded. Therefore, it is difficult to conclude that the recent genetic information on the NK isolate exactly reflects the past. Furthermore, due to the lack of information on the present isolates from North Korea, it is also very difficult to figure out the current distribution of the PvCSP gene variants in the Korean peninsula. To understand the PvCSP gene polymorphism, further investigation on more isolates in whole Korean peninsula is indispensible.
As shown in Fig. 5, the two genotypes of SK-A and SK-B can be easily distinguished. The DNA sequencing data and the RFLP pattern of the 30 specimens coincided very well (data not shown). Therefore, RFLP can be more useful in distinguishing the two genotypes of P. vivax found in South Korea instead of classifying them by using the gene sequencing technique.
In conclusion, the present findings suggest that two genotypes of P. vivax coexist in the epidemic area of South Korea and the isolates are from the East Asian group. Also, RFLP analysis other than the sequencing will be useful to study the PvCSP polymorphism of P vivax in South Korea.
Notes
This study was supported by a grant (HMP-96-M-2-1057) of Good Health R&D project, Ministry of Health and Welfare, Republic of Korea.