Cited By
Citations to this article as recorded by

East-to-west dispersal of bird-associated ixodid ticks in the northern Palaearctic: Review of already reported tick species according to longitudinal migratory avian hosts and first evidence on the genetic connectedness of Ixodes apronophorus between Sibe
Andor Pitó, Denis Fedorov, Vojtěch Brlík, Jenő Kontschán, Gergő Keve, Attila D. Sándor, Nóra Takács, Sándor Hornok
Current Research in Parasitology & Vector-Borne Diseases.2024; 6: 100201.
CrossRef Theileria and Babesia infection in cattle – First molecular survey in Kazakhstan
Marat Kuibagarov, Riza Makhamed, Assylbek Zhylkibayev, Maxat Berdikulov, Sarsenbay Abdrakhmanov, Mazhit Kozhabayev, Ilyas Akhmetollayev, Kasim Mukanov, Anara Ryskeldina, Yerlan Ramankulov, Alexandr Shustov, Christian Bauer, Alexandr Shevtsov
Ticks and Tick-borne Diseases.2023; 14(1): 102078.
CrossRef Spotted fever group rickettsiae in hard ticks in eastern and southern Kazakhstan
Qiaoyan Dong, Meihua Yang, Fengshi Li, Yuqing Jia, Kadyken Rizabek, Kenesbay Kairullayev, Otarbayev Bauyrzhan, Ketan Adil, Kazkhan Oralhazi, Yuanzhi Wang
Ticks and Tick-borne Diseases.2023; 14(6): 102238.
CrossRef Population genetic structure and demographic history of Dermacentor marginatus Sulzer, 1776 in Anatolia
Ömer Orkun, Eneshan Sarıkaya, Anıl Yılmaz, Mesut Yiğit, Zati Vatansever
First detection of Rickettsia aeschlimannii in Hyalomma marginatum in Tibet, China
Jun Jiao, Yonghui Yu, Peisheng He, Weiqiang Wan, Xuan OuYang, Bohai Wen, Yi Sun, Xiaolu Xiong
Morphological, distributional, and molecular study of the Phlebotomus sand flies of Jordan
Rami M. Mukbel, Dania A. Kanaan, Marwan M. Abu Halaweh, Zuhair S. Amr
Journal of Vector Ecology.2024;[Epub]
CrossRef
Abstract
To date, there is no report on the genetic diversity of ticks in these regions. A total of 370 representative ticks from the south and east regions of Kazakhstan (SERK) and Xinjiang Uygur Autonomous Region (XUAR) were selected for molecular comparison. A fragment of the mitochondrial cytochrome c oxidase subunit I (cox1) gene, ranging from 631 bp to 889 bp, was used to analyze genetic diversity among these ticks. Phylogenetic analyses indicated 7 tick species including Hyalomma asiaticum, Hyalomma detritum, Hyalomma anatolicum, Dermacentor marginatus, Rhipicephalus sanguineus, Rhipicephalus turanicus and Haemaphysalis erinacei from the SERK clustered together with conspecific ticks from the XUAR. The network diagram of haplotypes showed that i) Hy. asiaticum from Almaty and Kyzylorda Oblasts together with that from Yuli County of XUAR constituted haplogroup H-2, and the lineage from Chimkent City of South Kazakhstan was newly evolved; and ii) the R. turanicus ticks sampled in Israel, Almaty, South Kazakhstan, Usu City, Ulugqat and Baicheng Counties of XUAR were derivated from an old lineage in Alataw City of XUAR. These findings indicate that: i) Hy. asiaticum, R. turanicus and Ha. erinacei shared genetic similarities between the SERK and XUAR; and ii) Hy. marginatum and D. reticulatus show differences in their evolution.
Key words: Genetic diversity, hard tick, Kazakhstan, northwestern China
The Republic of Kazakhstan is located in Central Asia between 39°49°–55°49° N and 46°28°–87°18° E, with its western part extending into Eastern Europe. Kazakhstan is the ninth largest country in the world with a total area of 2,727,300 km
2, also ranking as the world’s largest landlocked country [
1,
2]. It borders Russia, Kyrgyzstan, Turkmenistan, Uzbekistan and China [
3]. Xinjiang Uygur Autonomous Region (XUAR), occupies 1/6 of China and borders 8 countries, covering an area of approximately 1,660,000 km
2 [
4]. The borderline between the south and east regions of Kazakhstan (SERK) and XUAR is 1,783 km long [
5]. The ecological environment, topography, climate and natural landscape are similar both in the SERK and XUAR [
6].
The mitochondrial cytochrome
c oxidase subunit I (
cox1) gene, also regarded as DNA barcoding, was informative in resolving recent diverging events between closely related species and at the intraspecific level [
7]. Recently, 14 tick species, including
Dermacentor nuttalli,
Dermacentor niveus,
Dermacentor marginatus,
Dermacentor silvaru,
Hyalomma asiaticum,
Hyalomma detritum,
Haemaphysalis punctata,
Haemaphysalis concinna,
Haemaphysalis erinacei,
Rhipicephalus turanicus,
Rhipicephalus sanguineus,
Argas vespertilionis,
Argas persicus, and
Ixodes kaiseri, have been molecularly characterized in the border regions of XUAR [
8]. At the same time, while at least 13 tick species have been identified in Kazakhstan, including
Dermacentor daghestanicus,
D. marginatus,
D. niveus,
Dermacentor reticulatus,
Hy. asiaticum,
Hyalomma anatolicum,
Hyalomma scupense,
Hyalomma marginatum,
Haemaphysalis punctata,
Haemaphysalis sulcata,
Rhipicephalus turanicus,
Boophilus calcaratus and
Ixodes persulcatus [
9,
10], and their molecular characteristics are still unknown. Here we report the genetic diversity of ticks in the SERK, in comparison with ticks from the XUAR.
During 2015–2019, a total of 4,392 hard ticks were collected from 605 domestic animals (287 sheep, 210 cattle, 101 camels, 7 dogs) and 16 hedgehogs at 24 sampling sites belonging to 15 districts of 5 Oblasts (East Kazakhstan, Almaty, Jambyl, South Kazakhstan and Kyzylorda) in the SERK. After morphological identification, 213 representative ticks were selected for further molecular analysis. Meanwhile, 157 representative ticks collected from marbled polecats, sheep and camels between 2015 and 2018 in 8 districts of the XUAR were used for comparison.
The genomic DNA was extracted from the selected 370 ticks individually using TIANamp Genomic DNA Kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. In this study, we methodologically omitted shorter than 450 bp
cox1 sequences, and chose 631–889 bp fragment to analyze inter- and intra-species genetic diversity. The primers used for PCR were LCO1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) [
11]. The cycling conditions consisted of an initial 5-min denaturation at 95°C, followed by 37 cycles at 94°C for 40 sec, 55°C for 40 sec, and 72°C for 1 min, with a final extension at 72°C for 10 min.
Sequences of the
cox1 were compared with GenBank data using BLAST program (
http://www.ncbi.nlm.nih.gov/BLAST/) after manual editing. This dataset was resampled 1,000 times to generate bootstrap values. Phylogenetic relationships were inferred using the Maximum Likelihood (ML) method. The best-fitting substitution model was determined with the Akaike Information Criterion using the ML model test implemented in MEGA 7.0 software [
12]. Genetic diversity was estimated as haplotype (h) and nucleotide diversity (π) indices with the programme DNAsp v5.10.1 [
13]. Median-joining (MJ) networks were generated with the software Network v5.10.1 [
14] to display the configuration of haplotypes [
15].
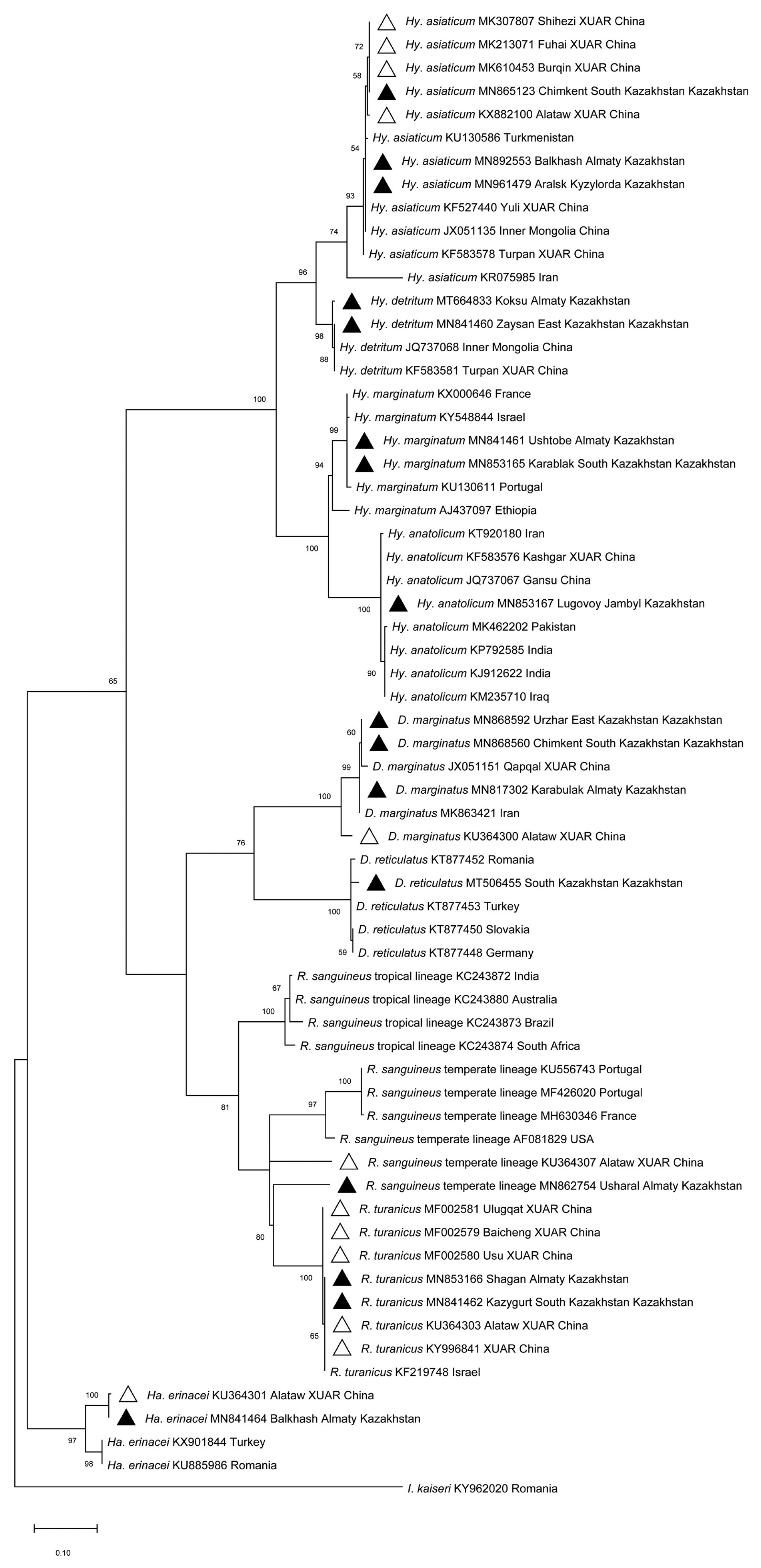
Morphological and molecular identification revealed the presence of 9 tick species in the 5 evaluated southeastern border oblasts of Kazakhstan, including Hy. asiaticum, Hy. detritum, Hy. anatolicum, Hy. marginatum, D. marginatus, D. reticulatus, R. sanguineus, R. turanicus and Ha. erinacei. Sequence identities ranged from 96.27% to 100% using BLAST analysis of the cox1 in intra-species. The sequences in this study were deposited in GenBank (Accession No.: MF002579-MF002581, MK213071, MK307807, MK610453, MN817302, MN841460-MN841462, MN841464, MN853165-MN853167, MN862754, MN865123, MN868560, MN868592, MN892553, MN961479, MT506455, MT664833, KU364300-KU364301, KU364303, KU364307, KX882100, KY996841).
Phylogenetic analysis based on the
cox1 (
Fig. 1) revealed that: i)
Hy. detritum ticks from East Kazakhstan Oblast (MN841460) clustered together with conspecific ticks from Turpan Region of XUAR (KF583581) and Inner Mongolia Province of China (JQ737068); ii)
Ha. erinacei ticks from Almaty Oblast, Kazakhstan (MN841464), and Altaw City of XUAR (KU364301) belonged to a different phylogenetic group than ticks from European countries; iii)
Hy. anatolicum ticks from Jambyl Oblast (MN853167), Karshgar City of XUAR (KF583576) and Iran (KT920180) represented an ancestral clade to ticks from Iraq, India and Pakistan; iv)
D. marginatus ticks from Qapqal County of XUAR (JX051151), East Kazakhstan Oblast (MN868592), and South Kazakhstan Oblast (MN868560) showed a separated branch with ticks from
D. marginatus in Alataw City of XUAR (KU364300) with high (100%) bootstrap support; and v)
R. sanguineus sensu lato from Alamaty Oblast (MN862754) was more closely related to a specimen from Iran than to ticks from Altaw City of XUAR (KU364307).
Haplotypes is defined as specific combinations of alleles on an individual chromosome [
16]. The network diagram of
Hy. asiaticum showed the haplotypes in the SERK and XUAR based on the
cox1. The analyses are as follows: i) the H-2 haplotype was the most dominant and the widespread in SERK and XUAR, such as Almaty Oblast (MN892553), Kyzylorda Oblast (MN961479) and Yuli County of XUAR (KF527400); ii) the H-5, H-6, and H-7 haplotypes were originated from the H-2 haplotype; and iii) the H-3 and H-4 haplotypes were furtherly originated from the H-6 haplotype. Interestingly, the H-1 haplotype, being distributed in South Kazakhstan, was derivated from the H-3 haplotype, which was distributed in Shihezi City, Fuhai County and Alataw City of XUAR. Meanwhile, 3 lineages of
R. turanicus from Almaty and South Kazakhstan Oblast (MN853166 and MN841462, H-1 haplotype), XUAR (MF002579-MF002581, H-3 haplotype), and Israel (KF219748, H-4 haplotype), originated from an ancestral lineage from Alataw City of XUAR (KU364303, H-2 haplotype) (
Fig. 2). In addition, our study revealed many highly divergent haplotypes for
Hy. asiaticum, e.g. H-1, H-4, H-7, and H-8, while many highly divergent haplotypes for
R. turanicus, e.g. H-1, H-3, and H-4, this phenomenon maybe be related to a long and complex evolutionary history of
Hy. asiaticum and
R. turanicus in the XUAR and SERK.
Based on ecological feature,
Hy. scupense is a one-host tick while
Hy. detritum is a 2-host species, although they are identical in morphological characteristics [
17]. Here
Hy. detritum ticks were all sampled from herbivorous livestock, and phylogenetic analyses showed that
Hy. detritum in East Kazakhstan Oblast (MN841460) shared a common branch with these in Turpan Region (the north of XUAR, KF583581) and Inner Mongolia Province of China (JQ737068), and that
Hy. detritum ticks in Almaty Oblast (MT664833) were their ancestral lineages.
The brown dog tick
R. sanguineus (Latreille, 1806), which is generally accepted as the taxonomic baseline for the
R. sanguineus group, is the most widely distributed tick species globally [
18]. At least 2 lineages of
R. sanguineus seno lato (s.l), namely temperate and tropical lineages, were presented [
19]. Here phylogenetic analysis indicated
R. sanguineus both Almaty Oblast in Kazakhstan (MN862754) and Alataw City of XUAR (KU364307) belonged to
R. sanguineus s.l temperate lineages, which was a separated clade with
R. sanguineus tropical lineage from India, Australia, Brazil and South Africa.
The mitochondrial
cox1, as the standard DNA barcode, is the best choice for identification of tick species [
7]. In the present study, 7 tick species from the SERK, including
Hy. asiaticum,
Hy. detritum,
Hy. anatolicum,
D. marginatus,
R. sanguineus,
R. turanicus and
Ha. erinacei, were clustered together with conspecific ticks from the XUAR. Nevertheless, the mitochondrial lineages were unknown in
Hy. marginatus and
D. reticulatus ticks from the SERK and XUAR, as for the
cox1 data of
Hy. marginatum and
D. reticulatus from the XUAR were absent. Animal transportation likely contributed to the dispersal of different haplotypes. Movements of wildlife and migratory birds are well-documented in these regions [
20], which might contribute to common genetic lineages of tick species. In the future, it is vital to sample more tick species both in the SERK and XUAR, which is helpful to explore their commonness and difference in tick’s evolution.