Genetic and Morphologic Identification of Spirometra ranarum in Myanmar
Article information
Abstract
In the present study, we identified a Spirometra species of Myanmar origin (plerocercoid) by molecular analysis using mitochondrial cox1 and nad1 genes, as well as by morphological observations of an adult tapeworm. Spargana specimens were collected from a paddy-field in Taik Kyi Township Tarkwa Village, Yangon, Myanmar in December 2017. A total of 5 spargana were obtained from 20 frogs Hoplobatrachus rugulosus; syn: Rana rugulosa (Wiegmann, 1834) or R. tigrina (Steindachner, 1867). The plerocercoids were used for experimental infection of a dog. After 4 weeks of infection, an adult tapeworm was recovered from the intestine of the dog. Morphologically, the distinct features of Spirometra sp. (Myanmar origin) relative to S. erinaceieuropaei and S. decipiens include a uterine morphology comprising posterior uterine coils that larger than the terminal uterine ball and coiling of the uteri diagonally (swirling) rather than spirally. The cox1 sequences (1,566 bp) of the Myanmar-origin Spirometra species showed 97.9% similarity to a reference sequence of S. decipiens (GenBank no. KJ599679) and 90.5% similarity to a reference sequence of S. erinaceieuropaei (GenBank no. KJ599680). Phylogenetic tree topologies were identical and presented high confidence level of values for the 3 major branches of the 3 Spirometra species in cox1 and nad1 genes. These results indicated that Myanmar-origin Spirometra species coincided with those of S. ranarum and may be considered as a valid species.
INTRODUCTION
The genus Spirometra belongs to the family Diphyllobothriidae and order Diphyllobothriidea and includes intestinal parasites of cats and dogs. Human sparganosis is a zoonotic disease caused by infection with the larval forms (procercoid/plerocercoid) of Spirometra species. The genus Spirometra has been described with morphological features of spirometrid species under the generic name Diphyllobothrium as found in China and including S. erinaceieuropaei (Rudolphi, 1819), S. decipiens (Diesing, 1850), S. ranarum (Gastaldi, 1854), S. mansoni (Cobbold, 1882) S. houghtoni (Syn. S. mansoni, Faust et al., 1929) and S. okumurai (Faust et al., 1929) by Faust et al. [1]. Spirometra species in North America have been identified such as S. mansonoides (Mueller, 1935) which possesses a characteristics C-shaped outer loop of the uterus [2]. Five Spirometra species, S. decipiens, S. mansoni, S. gracilis (Baer, 1927), S. longicollis (Parodi and Widakowich, 1917) and S. mansonoides have been reported from wild fields in South America [3].
Although human sparganosis is sporadically reported around the world, it is frequently reported in Asian countries. Among these species in the genus Spirometra, where S. erinaceieuropaei is known as the only Spirometra species in Asian countries including China, Indonesia, Japan, Korea and Thailand [4,5]. Many investigations of the spargana in frogs and snakes have been reported in field studies covering 27 Provinces in China and are critical for the eventual implementation of measures for the prevention and control of human sparganosis as more than 1,000 cases of human sparganosis have been reported in China [6]. The nucleotide sequence variations of spargana in frogs (Rana nigromaculata) and snakes were 0–8.4% for cox1, 0–1.5% for cox3, 0–2.8% for nad1, and 0–2.7% for nad4 in Hunan, Guangxi, and Henan Provinces, China, respectively [6–10]. In Thailand, 55 human sparganosis were reported as 53 non-proliferative and 2 proliferative. Among them, 12 cases were identified as infection with S. erinaceieuropaei plerocercoids by PCR-based molecular identification of the worms [11]. Intraspecies variation of cox1 gene of S. erinaceieuropaei was reported from Japan, India and Indonesia by Okamoto et al. (<2.6%) [12]. Molecular identification of Spirometra species collected from animals has been studied using partial sequence analysis of the cox1 gene of spargana isolated from frogs (Hoplobatrachus rugulosus) and snakes (Ptyas korros) from Lao PDR, Myanmar and Thailand [13]. The results revealed that nucleotide variation in the partial cox1 sequence (429 bp) among the spargana ranged from 0–3.5%. Those molecular studies have indicated that 2 more genotypes were found in Spirometra species in those endemic regions revealing genetic variation of mitochondrial DNA sequences. As with the above studies, the most important causative agent of human sparganosis is S. erinaceieuropaei in Asian countries. Two Spirometra species currently being considered as valid are S. erinaceieuropaei and S. decipiens by morphological and molecular approaches using complete mitochondrial genome analysis [14]. Therefore, identification of Spirometra species requires the use of molecular and morphological techniques to better understand the epidemiological status of Spirometra spp. in its endemic regions including Asian countries.
One Spirometra species in Myanmar is known as S. ranarum, which was first reported by Gastaldi (1854) from Rana esculenta (syn: Pelophylax esculentus) in Italy, and Meggitt described it as S. ranarum from a dog fed spargana isolated from the same frog host by Gastaldi (1854) in Myanmar [15]. In the present study, we identified a Spirometra species of Myanmar origin (plerocercoid) by molecular analysis using 2 complete mitochondrial genes, cytochrome c oxidase I (cox1) and NADH dehydrogenase subunit 1 (nad1), as well as by morphological observations of an adult tapeworm experimentally obtained from a dog fed with the plerocercoid.
MATERIAS AND METHODS
Specimens
Spargana specimens were collected from a paddy-field in Taik Kyi Township Tarkwa Village, Yangon, Myanmar in December 2017. A total of 5 spargana were obtained from 20 frogs (Hoplobatrachus rugulosus; syn: Rana rugulosa [Wiegmann, 1834] or R. tigrina [Steindachner, 1867], commonly known as Chinese edible frog, East Asian bullfrog or Taiwanese frog). The 5 plerocercoids were then used to infect a dog. After 4 weeks of infection, an adult tapeworm was recovered from the intestine of the dog. The collected worm was fixed with 10% neutral buffered formalin before it was placed in hot water (90°C) for 5 min and microscopic observations were then performed following carmine staining. The vaginal opening, uterus, uterine pore, cirrus, genital pore, testes, and vitellaria were observed with carmine-stained mature and gravid proglottids according to the methods of Meggitt and Faust et al. [1,15]. Gravid proglottids in longitudinal section were stained with hematoxylin and eosin for observing genital organs. Some of the gravid proglottids were preserved in a −70°C deep freezer and in 70% ethanol for molecular studies. The experimental animal was treated according to the recommendations of the Guide for Institutional Animal Care and Use Committees (IACUC) of Chungbuk National University, Cheongju, Korea (Permission No. CBNUA-1090-18-02). Parasite materials used in this study were provided by the Parasite Resource Bank of the Korea National Research Resource Center, Republic of Korea (PRB000720).
PCR and DNA sequencing
Total genomic DNA was extracted from a single proglottid using a DNeasy tissue kit (Qiagen, Valencia, California, USA) according to the manufacturer’s instructions. The primers were used to amplify the mitochondrial cox1 gene (Spi-CO1F: 5′-GAC TAA GTG TTT TCA AAA CAC TAA GTG -3′ and Spi-CO1R: 5′CAC CCT ACC CCT GAT TTA CAA AAT-3′) and nad1 gene (Spi-ND1F: 5′-GGA GAA TAT TGG TTT GTC TAA CCA-3′ and Spi-ND1R: 5′-CCT TCT TAA CGT TAA CAG CAT TAC GAT-3′). PCR was performed in a 50 μl of reaction volume containing 50 ng of genomic DNA, 10 pmol of each primer, 5 μl of 10X buffer, 10 mM dNTP mix, 25 mM MgCl2, and 2.5 units Taq polymerase (High Fidelity PCR system, Roche, Mannheim, Germany). PCR was employed using a GeneAmp PCR system 9700 instrument (Applied Biosystems, Langen, Germany) under conditions comprising: 94°C for 3 min; 35 cycles of 94°C for 30 sec, 45°C for 40 sec, 72°C for 60 sec, and a final step at 72°C for 5 min. The cox1 and nad1 DNA fragments were extracted using a QIAquick PCR purification kit (Qiagen). DNA sequencing was performed by a private company (COSMOGENETECH, Daejeon, Korea). Cycle sequencing was performed using a Big-Dye terminator kit (version 3.1, Applied Biosystems, Foster City, California, USA). Reaction products were sequenced directly using a DNA sequencer (ABI3730XL, Applied Biosystems).
DNA sequence analyses
DNA sequences were assembled and aligned using the Geneious 9.0 software program (Biometer, Auckland, New Zealand). These sequences were identified by comparison with the sequences of S. erinaceieuropaei and S. decipiens in the Gen-Bank database. Phylogenetic analyses were evaluated using Bayesian inference (BI) and maximum-likelihood (ML). ML analyses of cox1 and nad1 used RAxML v. 7.3.1 [16] after GTR+ I+G substitution model sampling was chosen according to the Modeltest using the program Partition Finder [17]. BI analyses were used in MrBayes 3.2 and running four simultaneous Monte Carlo Markov chains (MCMC) for 10 million generations, sampling every 1,000 generations and discarding the first 25% generations as burn-in [18]. Phylogenetic trees were constructed using full mitochondrial cox1 (1,566 bp) and nad1 (891 bp) sequences of S. erinaceieuropaei (KJ599680), S. decipiens (KJ599679), D. nihonkaiense (EF420138) and D. latum (DQ985706). Pairwise sequence differences based on the full mitochondrial cox1 and nad1 sequences were analyzed using the NJ method with the K2P substitution model.
RESULTS
Sequence similarity and phylogenetic relationships
The cox1 sequences (1,566 bp) of the Myanmar-origin Spirometra species showed 97.9% similarity to reference sequences of S. decipiens (GenBank no. KJ599679) and 90.5% similarity to a reference sequence of S. erinaceieuropaei (GenBank no. KJ599680). The similarity to other Diphyllobothrium species was 79.5% (D. nihonkaiense) and 80.1% (D. latum). The similarity of nad1 sequences (891 bp) from Myanmar-origin Spirometra species to the reference sequence was 97.8% (S. decipiens), 89.1% (S. erinaceieuropaei), 73.8% (D. nihonkaiense), and 73.3% (D. latum) (Table 1).

Percentage pairwise sequence homologies of mitochondrial cox1 and nad1 genes of Spirometra species in Myanmar and various Spirometra species, Diphyllobothrium latum and D. nihonkaiense
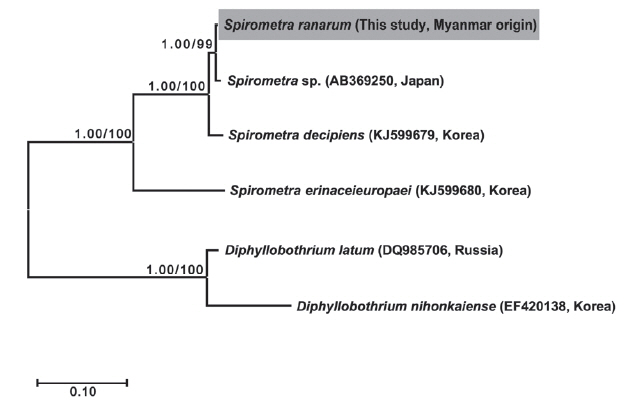
Phylogenetic analyses of Spirometra species were conducted using the 2 methods (BI and ML) based on the cox1 and nad1 genes of S. decipiens, S. erinaceieuropaei, D. nihonkaiense, and D. latum. The cox1 sequences (1,566 bp) revealed 33 polymorphic sites, with 26 synonymous and 7 non-synonymous substitutions, while nad1 sequences (891 bp) showed 19 polymorphic sites, with 16 synonymous and 3 non-synonymous substitutions between the Myanmar-origin Spirometra species and S. decipiens (GenBank no. KJ599679). Phylogenetic tree topologies generated using the 2 analytic methods were identical and presented high level of confidence values for the 3 major branches of the 3 Spirometra species in the cox1 and nad1 genes (Fig. 1).
Morphological characteristics
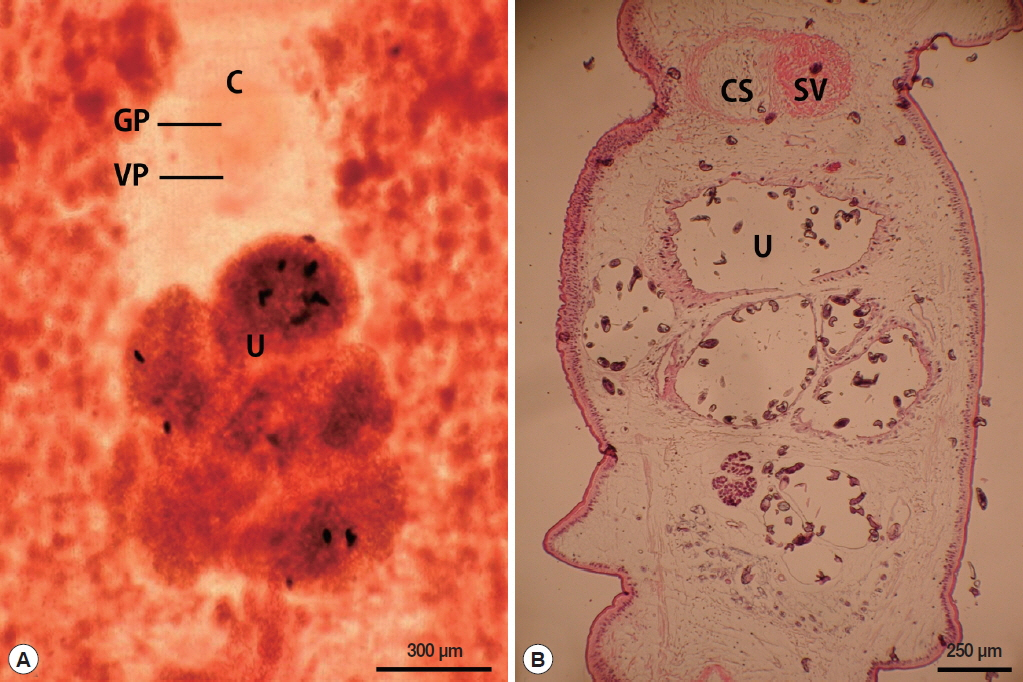
The length of the whole strobila was 237 cm. The mature proglottids with eggs were first visible almost 32 cm downward from the neck. The average width and length of the gravid proglottid was 15.0 mm by 2.0 mm (n=10) (Table 2). The genital and vaginal pore were closely situated ventrally on the midline in the anterior 1/3 of the segment. The uterine pore was on the midline behind the anterior margin of the terminal ball 370–420 μm in diameter (n=10). The uterus consisted of 3 loops with a diagonal direction of the second turn (Fig. 2A). A longitudinal section of the cirrus sac showed an oval structure 175–250 μm in diameter (n=10). The seminal vesicle was almost round in shape 197–222 μm in diameter. The testes (n=10) measured an average 74 μm in width. The eggs measured 52–55 μm by 31–35 μm in transverse diameter (n=10). The ovary was highly dendritic and was situated near the posterior margin of the proglottids (Fig. 2B).
DISSCUSSION
The larval stage (plerocercoid) of S. ranarum was first described by Gastaldi (1854) from Rana esculenta (syn: Pelophylax esculentus) from Italy. In 1924, Meggitt reported spargana from frogs (Rana tigrina) from Yangon, Myanmar, and autopsies of dogs fed some of the spargana revealed the presence of adult spirometrids [15]. Meggitt, Joyeux and Baer (1927) described in detail this species with complete life cycle through intermediate hosts found and showed it to be suitable for final hosts [1]. Faust et al. (1929) found this species from natural infections of cats and dogs in Beijing, Xiamen, Canton and by experimental feeding of spargana obtained from dogs in Fujian [1]. The morphological characteristics of S. ranarum that have been detailed included the following features: 3–4½ uterine coils of which the posterior 2 are broader and more dilated than the terminal uterine ball, highly dendritic ovaries, the testes and vitellaria are closely positioned to the lateral margins of the lower 2 coils of the outer uterus, the vaginal and male genital pores are located in the median plane at small a distance anterior to the terminal uterine coil, and the eggs are similar in shape to those of S. decipiens [1].
The morphological features of the spirometrid tapeworm observed in the present study resembled that of previous studies by showing the genital and vaginal pore closely situated ventrally on the midline at the anterior 1/3 of the segment. The uterine pore was on the midline behind the anterior margin of the terminal uterine ball. The uterus consisted of 3 loops with a diagonal direction of the second turn and these 2 uterine coils were larger than the terminal uterine ball. The distinct features of S. ranarum relative to other Spirometra species include a uterine morphology comprising posterior uterine coils that are larger than the terminal uterine ball and coiling of the uteri diagonally (swirling) rather than spirally. These morphological characteristics of the Spirometra specimen (Myanmar origin) examined in this study are consistent with this S. ranarum specimen in terms of shape of the uterus.
Molecular approaches for taxonomy are usually based on genetic markers for identification and are applied to genetic variations especially among a group of morphologically similar parasites. The mitochondrial DNA sequences of cox1, cox3, nad3, and nad4 genes have been used as genetic markers for phylogenetic reconstruction, taxonomic identification and epidemiological investigations [6–14]. Recent molecular data of Spirometra species reported from Asian, South American and African countries show that there are at least 4 Spirometra species in those endemic areas [3]. The nucleotide sequence variation of S. erinaceieuropaei ranged from 0.0–3.5% in China, Myanmar, Thailand and in Lao PDR as determined from frogs and snakes. Some studies from Japan, India and Indonesia found 2.6% variation in the DNA sequence of cox1 genes obtained from dogs [12]. The degree of mtDNA sequence divergence of cytochrome b (cob) gene between sister or congeneric species, and con-familial genera was greater than 2% in amphibian, reptilian, avian and mammalian species [19]. The closely related species of vertebrates showed more than 2% of sequence divergence in the cox1 gene [20]. In this study, sequence differences in cox1 and nad1 genes between Spirometra species (Myanmar origin) and S. decipiens was 2.1%, while those of S. erinaceieuroapei were 9.5% (cox1) and 11.9% (nad1), respectively. Therefore, the results indicated that the examined Spirometra species from Myanmar matched S. ranarum in terms of morphological characteristics and mitochondrial sequence divergence. Some previous studies have reported that sequence divergence of the partial cox1 gene (300–450 bp) in Spirometra species was greater than 3.0%, whereas sequence differences in the full mitochondrial cox1 sequence was 2.1%. The results of this comparative study indicated that analysis of short sequences of the cox1 gene may give rise to bias, and that analysis of the complete sequence data provides more reliable data.
Regardless of the molecular and phylogenetic results, the epidemiological status of Spirometra species should be considered in terms of species distribution and human sparganosis rather than assessment based primarily on GenBank DNA sequences. The Asian isolates of Spirometra species were revealed to be S. decipiens and S. ranarum in Myanmar, Thailand, Lao PDR, China, Korea and Japan, whereas only S. erinaceieuropaei human cases were found in Korea and Japan. These findings indicated that S. erinaceieuropaei, S. decipiens, and S. ranarum are a possible cause of human sparganosis. Further studies are needed to better understand the taxonomic and epidemiological status of Spirometra species for investigations of African and South American isolates.
This study has demonstrated the identification of Myanmarorigin Spirometra species by mitochondrial cox1 and nad1 sequence analysis and morphological examination of the adult worm. Furthermore, these results may indicate that S. ranarum is a valid species.
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation of Korea (no. 2017R1D1A3B03035967). Materials used were provided by the Parasite Resource Bank of Korea (PRB000720).
Notes
CONFLICT OF INTEREST
We have no conflict of interest related to this work.