Molecular and biochemical characterization of a novel actin bundling protein in Acanthamoeba
Article information
Abstract
Actin binding proteins play key roles in cell structure and movement particularly as regulators of the assembly, stability and localization of actin filaments in the cytoplasm. In the present study, a cDNA clone encoding an actin bundling protein named as AhABP was isolated from Acanthamoeba healyi, a causative agent of granulomatous amebic encephalitis. This clone exhibited high similarity with genes of Physarum polycephalum and Dictyostelium discoideum, which encode actin bundling proteins. Domain search analysis revealed the presence of essential conserved regions, i.e., an active actin binding site and 2 putative calcium binding EF-hands. Transfected amoeba cells demonstrated that AhABP is primarily localized in phagocytic cups, peripheral edges, pseudopods, and in cortical cytoplasm where actins are most abundant. Moreover, AhABP after the deletion of essential regions formed ellipsoidal inclusions within transfected cells. High-speed co-sedimentation assays revealed that AhABP directly interacted with actin in the presence of up to 10 µM of calcium. Under the electron microscope, thick parallel bundles were formed by full length AhABP, in contrast to the thin actin bundles formed by constructs with deletion sites. In the light of these results, we conclude that AhABP is a novel actin bundling protein that is importantly associated with actin filaments in the cytoplasm.
INTRODUCTION
The actin cytoskeletons of eukaryotic cells are dynamic meshworks and are involved in many biological phenomena, such as cell motility, cell adhesion, intracellular transport, endo- and exocytosis, cytokinesis, and cell morphology (Delanote et al., 2005). In order to accomplish these numerous functions and respond rapidly to internal and external stimuli, a large number of accessory proteins are also engaged (Winder, 2003). Among these, the actin bundling proteins are responsible for the assembly of microfilaments in non-muscle cells by crosslinking filaments into parallel arrays, and thereby, provide mechanical support to the cytoplasm and reinforce cellular protrusions (Tseng et al., 2002). Actin bundling proteins are characterized by the presence of multiple actin binding sites per monomer, and possess domains which are responsible for various controlling and structural functions. These regulatory domains include phosphorylation sites, structural repeated domains, and EF-calcium binding hands (Bartles, 2000). To date, 13 actin binding proteins that cross-link actin filaments have been identified from among 162 distinct and separate actin binding proteins in various organisms (dos Remedios et al., 2003).
Members of the Acanthamoeba are considered model organisms, and have increased our understanding of eukaryotic cytoskeletal processes. A. castellanii, one of the most intensely investigated non-muscle cells (Ueno and Korn, 1986), has been widely used to study the molecular mechanisms of actin polymerization during amoeboid locomotion (Pollard and Ostap, 1996), because of the organism's well developed cytoskeleton and the presence of actin in abundance (Gordon et al., 1976). Moreover, actin binding proteins, such as, actophorin (Maciver et al., 1991), actobindin (Lambooy and Korn, 1986), 23,000, 28,000, 32,000 and 38,000 Da gelation proteins (Maruta and Korn, 1977), and a calcium-sensitive actin gelation protein (Pollard, 1981) in Acanthamoeba have been purified and characterized.
In this study, we identified and characterized a cDNA clone encoding a 32 kDa actin bundling protein isolated from Acanthamoeba, which we name AhABP. AhABP was characterized by sequence analysis, intracellular localization, and by actin binding analyses
MATERIALS AND METHODS
Amoebae cultivation
A. healyi (ATCC#30886) trophozoites, originally isolated from a case of GAE (Moura et al., 1992), were obtained from the American Type Culture Collection (Rockville, Maryland, USA) and were grown axenically at 25°C in PYG (peptone-yeast-glucose) medium (10 g proteose peptone, 10 g yeast extract, 10 ml - 50% glucose, 10 ml - 0.5 M Na2HPO4, 10 ml - 0.5 M K2HPO4 in 970 ml glass distilled water with the final pH adjusted to 6.5) at 25°C incubator (Sanyo, San Diego, California, USA).
Sequence analysis
The cDNA used in this study was derived from a previously constructed cDNA library of A. healyi (Kong et al., 2001). Sequences obtained were searched for homology using the Basic Local Alignment Tool X (BLASTX) program at the National Institute for Health. Predicted motifs and secondary structures were obtained from Predictprotein, at the Columbia University Bioinformatics Center. The molecular mass and isoelectric point of AhABP were computed from deduced amino acid residues using the Translate program at the ExPasy Proteomics Server. Protein families and domains were identified using ProSite at the same server. Phylogeny was obtained by aligning actin bundling proteins using CLUSTALX version 1.82 with a low gap penalty.
Northern blot analysis
Total RNA was isolated from trophozoites of A. healyi OC-3A, A. castellanii Castellani and A. castellanii Neff. The probe, a 389 bp sized PCR fragment of the AhABP gene for actin bundling protein in Acanthamoeba healyi (AhABP), was randomly primed with a DIG-dUTP (Roche, Manheim, Germany). Hybridization of membranes was performed using ExpressHyb Hybridization Solution (Clontech, San Jose, California, USA), according to the manufacturer's instructions. Also, a 300 bp fragment of 18s rDNA from Acanthamoeba was derived by PCR method and was used as a loading standard for Northern blot expression levels. The membrane was deprobed and rehybridized with the probe of 18s rDNA as a RNA loading control.
Construction of expression vectors
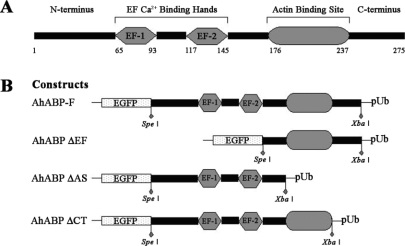
Transfection vectors for transient expression were constructed by ligating AhABP gene into pUb-EGFP expression vector containing an Acanthamoeba ubiquitin promoter (Kong and Pollard, 2002). Figure 1 shows the different expression vectors used in this study, including full-length AhABP cDNA (AhABP-F) and 3 deletion mutants corresponding to the fragments with deleted EF-hands (AhABP ΔEF), deleted actin binding site (AhABP ΔAS), and deleted C-terminus (AhABP ΔCT). Primers including Spe I at the 5' end and an Xba I at the 3' end were designed to amplify cDNA fragments, which were then inserted into pUb-EGFP expression vector behind EGFP.
Transfection
A. healyi grown to mid log phase were washed with PBS and then resuspended in PYG culture medium. Approximately 4 × 105 cells per well were seeded in a six-well culture plate in 3 ml of PYG medium and incubated overnight at 25°C. At least 4 µg of plasmid DNA was added to PYG medium to a volume of 100 µl, and 20 µl of Superfect transfection reagent (Qiagen, Hilden, Germany) was then added. After incubation for 10 min, 900 µl of PYG medium was added to reaction tubes containing the transfection complexes and this was then pipetted dropwise into the wells. Cells were incubated for 2-3 hr at 25°C, medium was removed, and cells were washed once with PBS. The cells in growth medium were incubated for 24-48 hr at 25°C. Transfection efficiencies were calculated by measuring the percentage of fluorescent cells among transfected cells using a CellQuest 3.2 FACScan (Beckton-Dickinson, Franklin lakes, New Jersey, USA).
Microscopy
Amoeba expressing green fluorescent protein were selected and allowed to adhere to a cell culture dish (BD Falcon, Franklin lakes, New Jersey, USA). The cells were observed under an Olympus IX70 fluorescent microscope equipped with a cooled CCD camera (Roper Scientific, Tucson, Arizona, USA). Alternatively, transfected amoebae were flattened with an agar overlay to observe intracellular structures in detail (Yumura et al., 1984). EGFP fluorescence was achieved using a 500-530 nm band pass filter. Images and time lapse photographs were acquired and analyzed using the Metamorph imaging system (Universal Imaging Corp., West Chester, Pennsylvania, USA).
Actin filament co-sedimentation assays
The actin bundling abilities of recombinant AhABPs were determined using actin-filament cosedimentation assays. AhABP-F and truncated versions of the protein were cleared at 4°C for 30 min at 37,000 × g in a centrifuge (Centrikon, Kontron, Italy) to remove contaminating proteins. High speed actin co-sedimentation assays were performed as previously described (Lim and Fechheimer, 1997). Briefly, 0.2 mM of AhABP-F or each of the truncated constructs and 24 mM of actin (Sigma, Steinheim, Germany) were mixed in a polymerization buffer composed of 20 mM PIPES, pH 7, 50 mM KCl, 50 µM MgCl2, 1 mM ATP, 0.2 mM DTT, 5 mM EGTA and 0.2 mM CaCl2 (low conc.) or 5 mM (high conc.) to a final volume of 200 µl. These AhABP/actin mixtures were then incubated for 24 hr at 4°C to allow polymerization and were then centrifuged at 250,000 × g (Beckman TLX-100, Ramsey, Minnesota, USA) for 1 hr at 4°C. Supernatant and pellet fractions were separated and the proteins in each fraction were analyzed by SDS-PAGE. AhABP and actin alone were also centrifuged using the same conditions and loaded as controls.
Transmission electron microscopic examination of actin bundles
Negative staining of mixtures of actin and AhABPs was performed as previously described (Fechheimer and Furukawa, 1993). The same amounts of AhABP-Full and actin as used in co-sedimentation assays were prepared and allowed to react in polymerization buffer containing 5 mM CaCl2. After incubation at 4°C for overnight, a mixture was adsorbed onto a 300-mesh copper grid coated with 0.3% Formvar and carbon for 5 min. The grid was then washed and then stained with 2% uranyl acetate for 2 min. The actin bundles formed by AhABP were visualized under a transmission electron microscope (Hitachi-H 7100, Tokyo, Japan)
RESULTS
Sequence analysis
The cDNA clone found to be homologous to actin bundling protein was verified to contain a single open reading frame gene consisting of 828 bp. The deduced amino acid sequence of this gene revealed a protein of 275 amino acid residues with a predicted molecular mass of 32,000 Da and an isoelectric point of 5.79. The clone represented the entire coding sequence including the 3' termination codon, which was followed by presumably untranslated sequences and a poly A tail. Based on the results of a homology search, the gene was found to exhibit high similarity of up to 72% with actin bundling proteins of Physarum polycephalum and Dictyostelium discoideum. Phylogenetic analysis demonstrated that AhABP is more closely related to the ABP-46 P. polycephalum and the 34 kDa actin-bundling protein of D. discoideum rather than the actin bundling proteins of other organisms (Fig. 2). AhABP exhibits the characteristic features of actin binding proteins, i.e., phosphorylation sites, 2 structural repeated regions (amino acids 2-39 and 134-167), and 2 putative calcium-binding EF-hands (Fig. 3). Sspro (Pollastri and McLysaght, 2005) predictions of secondary structure indicated a high probability for the occurrence of helixes throughout the sequence and using the program Coils (Lupas, 1997), a 30-amino acid residue (EIRAARLALEEVNKSIKAYEAERYRLTEES) was identified in a coiled-coil domain. Analysis by Prosite (Falquet et al., 2002) of the deduced amino acid sequence found potentially important sites that may transduce information for intracellular signaling pathways, including N-myristoylation, tyrosine kinase phosphorylation, protein kinase C phosphorylation, and casein kinase II phosphorylation.
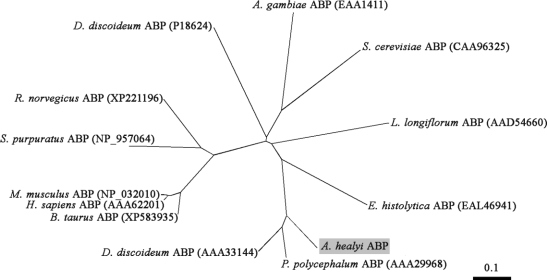
Phylogenetic tree derived by aligning AhABP with other actin bundling proteins in the database. The AhABP gene was found to be 72% and 70% identical with the ABP-46 P. polycephalum and the 34 kDa ABP D. discoideum respectively. Accession numbers are indicated in parenthesis.
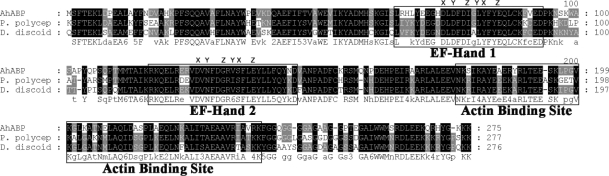
Alignment of AhABP with other actin bundling proteins identified from P. polycephalum and D. discoideum. Two regions of the amino acid sequence homologous to EF-hand calcium binding sites and a segment homologous to actin binding site of Dictyostelium 34 kDa protein are illustrated. The coordinates for Ca2+ ion binding in EF hands are indicated as x, y, z, -y, -x and -z.
Presence of EF-hands
AhABP was found to contain 2-centrally located EF-hand Ca2+ binding sites interposed by a 20-amino acid residue. The predicted calcium binding hands exist as a pair and contain the helix-loop-helix structure sometimes called the calcium-binding loop, which can be recognized by the presence of a consensus sequence. Alignment of EF-hands revealed that the AhABP loops contain the essential sites to bind the calcium atom coordinates x, y, z, -y, -x and z (Fig. 3).
Northern blot analysis
Northern blot analysis was performed to determine the expression of AhABP in different strains of Acanthamoeba, namely A. healyi OC-3A, A. castellanii Neff, and A. castellanii Castellani. Total RNA from these 3 strains showed a single band of approximately 0.8 kb corresponding to the size of the cDNA clone, and thus supported the notion that the cDNA clone is of full length. All Acanthamoeba strains showed detectable levels of AhABP mRNAs, with highest expression in A. healyi (Fig. 4).

A. Northern blot analysis of the AhABP gene in various Acanthamoeba strains. Lanes: 1, A. healyi OC-3A; 2, A. castellanii Neff and 3, A. castellanii Castellani. A single band of 0.8 kb corresponds to the size of the cDNA indicating that the clone was of full length. B. The expression level 18s rDNA was used as a loading standard.
Localization and distribution
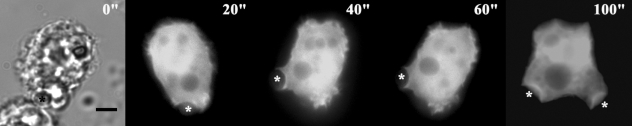
The transfection efficiencies of the constructs ranged from 0.5 to 3%. Transfection results showed that EGFP-AhABP-F was localized in cortical cytoplasm, excluding the nucleus and vesicles, but was also present at cell peripheries and in pseudopods (Fig. 5). Fig. 6 shows the localization of AhABP-F as observed in cells undergoing phagocytosis after adding heat-killed yeast cells to transfected amoeba in a microwell dish. EGFP fused AhABP-F accumulated in regions that came in contact with yeast cells and did not diminish even after completely enclosing yeast cells, indicating the presence of AhABP in mature phagosomes. Fig. 7 shows the formation of ellipsoidal inclusions at cellular peripheries expressing AhABP ΔEF, AhABP ΔAS, and AhABP ΔCT. In addition to the accumulation of these aggregates, most cells expressing site-deleted fragments exhibited rounding and membrane blebbing. These results suggest that the regulatory roles of EF-hands, the actin binding site, and the C-terminus are necessary for the proper binding of actin filaments by AhABP. The appearance of these aggregations was not manifested in cells expressing AhABP-F.
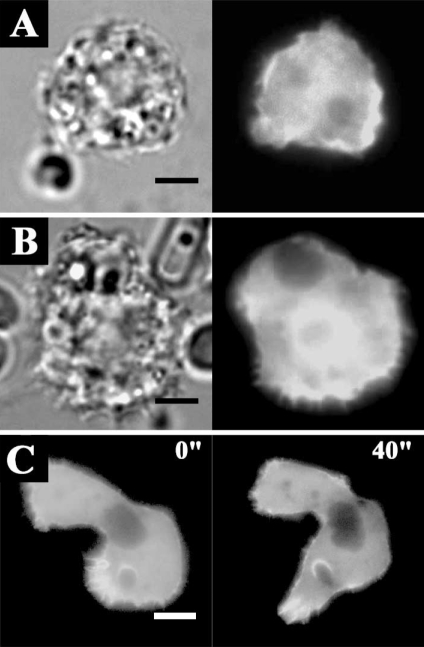
Fluorescence micrographs of A. healyi expressing full length AhABP fused with EGFP. Results of transfection into live amoeba cells indicated that AhABP was localized to cell (A) peripheries and (B) pseudopods. Bars: 10 µm.
Fluorescence micrographs of A. healyi expressing full-length AhABP-EGFP showing that the protein is present in mature phagosomes. White asterisks indicate the positions of yeast cells during phagocytosis. Bar: 10 µm.
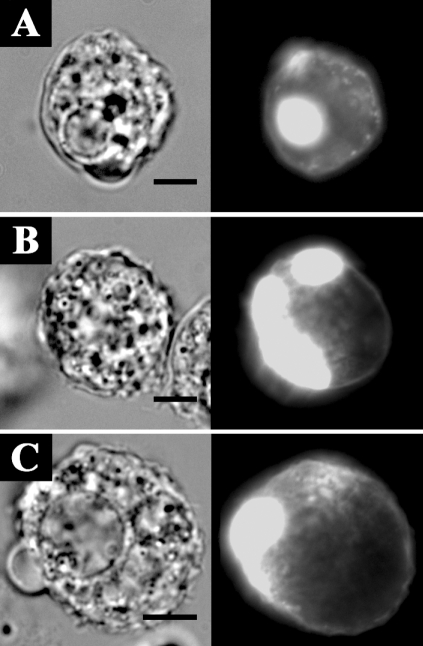
Localization and distribution of EGFP fused AhABP-deletion mutants in transfected cells. Different constructs, i.e., (A) AhABP ΔEF, (B) AhABP ΔAS, and (C) AhABP ΔCT were generated to investigate the relation between these regions and actin binding. Cells expressing these fragments demonstrated the formation of large aggregates located at cell peripheries. Bars: 10 µm.
Purification and expression of recombinant proteins
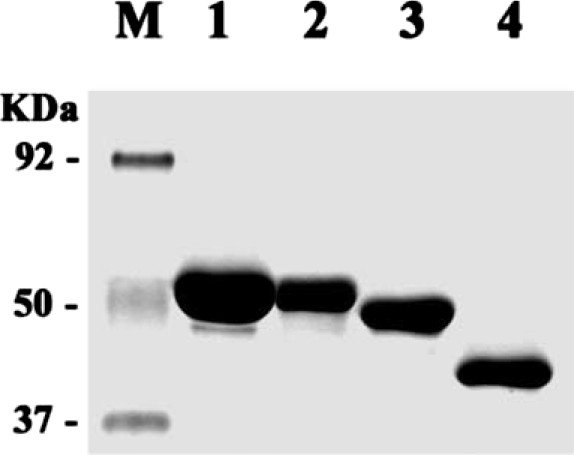
Thick bands of the fusion proteins, i.e., 58 kDa for AhABP-F-GST, 51 kDa for AhABP ΔCT-GST, 42 kDa for AhABP ΔAS-GST and 40 kDa for AhABP ΔEF-GST were detected (Fig. 8). AhABP-F and AhABP ΔEF were contained in the soluble fraction, whereas AhABP ΔCT and AhABP ΔAS were insoluble. Truncation of the C-terminal and deletion of the actin binding site may have caused these GST-fusion proteins to become unstable and insoluble.
Actin binding assay by co-sedimentation
Using the same conditions used for the co-sedimentation assay of Dictyostelium 34 kDa protein. Approximately 10 µM of free calcium is present in the polymerization buffer containing 5 mM CaCl2 as described in the Co-sedimentation assay of Dictyostelium (Fechhermer, 1987). As shown in Fig. 9A, recombinant AhABP wild type bound actin in the presence of high (5 mM) and low (0.2 mM) concentration of calcium by co-sedimentation assays, whereas constructs with deleted regions exhibited various binding degrees. For ΔEF and ΔCT, some of the actin remained in the supernatant, whereas in ΔAS, both the fusion protein and actin were present in the supernatant fraction at both low and high calcium concentrations. AhABP-F alone and actin alone also sedimented under the same conditions and were loaded as standards. AhABP-F or actin alone remained in the supernatant fraction and were not sedimented, as shown in Fig. 9B.
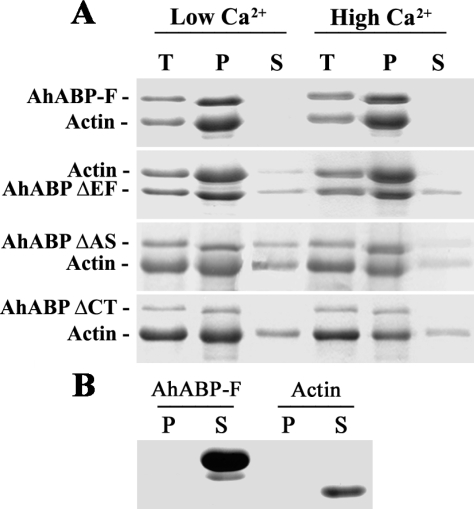
Co-sedimentation assay of AhABP-F and corresponding deletion mutants, namely, AhABP ΔEF, AhABP ΔAS and AhABP ΔCT. A. Mixtures of actin and AhABP-GST fusion proteins were allowed to polymerize in the presence of 0.2 mM (low conc.) or 5 mM (high conc.) CaCl2 and then centrifuged at 250,000 × g. B. AhABP-F alone and actin alone centrifuged under the same conditions did not sediment in the pellet obtained. Supernatants and pellets were separated and analyzed by SDS-PAGE. Lanes: T, total; P, pellet; S, supernatant.
Visualization of actin bundles by electron microscopy
To visualize the interaction between AhABP-F and AhABP deletion constructs, mixtures of AhABP and actin were allowed to copolymerize and examined by negative staining electron microscopy (Fig. 10). When the recombinant protein expressing AhABP-F was mixed with actin, actin was organized into large parallel arrays about 0.38 µm thick. The fragments, AhABP ΔEF and AhABP ΔCT formed thinner bundles, suggesting that these deleted sites are important for the formation of parallel actin bundles. Similar result was observe for AhABP ΔAS since the elimination of the actin binding site strongly reduced, but did not completely abolish actin bundling activity, as was indicated by the formation of thin actin filament bundles. These results are in agreement with the binding abilities observed in co-sedimentation assays.

Electron micrographs of AhABP-F and corresponding constructs. Mixtures of AhABP and actin in the presence of 5 mM CaCl2 viewed under a transmission electron microscope using negative staining: (A) AhABP-F, (B) AhABP ΔEF, (C) AhABP ΔAS and (D) AhABP ΔCT. AhABP-F showed the formation of thick parallel actin bundles, whereas the 3 constructs with deleted sites formed thinner actin filament. Bars: 0.2 µm.
DISCUSSION
In this study, we characterized an actin bundling protein from A. healyi, which we name AhABP. The predicted secondary structures of the AhABP sequence revealed a high probability for the occurrence of α-helices, and for the presence of glycosylation and phosphorylation sites, which are involved in transport and folding processes, and in the establishment of spatial boundaries in developing tissues. Also, the presence of 2 coiled-coil structures and abundant leucine in the C-terminus of AhABP may facilitate the dimerization of the protein.
Northern blot analysis showed that AhABP was more highly expressed in A. healyi OC-3A than in A. castellanii Neff and A. castellanii Castellani. Although no direct evidence supported that AhABP is related with Acanthamoeba pathogenicity, the enrichment of AhABP in dynamic areas of the cell suggests its association with areas rich in actin cytoskeleton. This indicates greater AhABP activity for processes that require actin, such as, membrane deformation leading to pseudopod protrusion, which is important for phagocytosis and cell motility. The expression of pathogenic activity requires a dynamic cytoskeleton that allows rapid movement, tissue penetration, and changes in parasite morphology (Voight et al., 1999). AhCoronin, a coronin isolated from A. healyi, exhibited similar gene expression patterns (Baldo et al., 2005). Several reports have been issued regarding the importance of actin binding proteins in cells. In the case of Entamoeba histolytica, myosin IB is highly concentrated in phagosomes, and enables this parasite to phagocytose human epithelial cells, immune cells, and erythrocytes (Voight et al., 1999).
The intracellular localization of AhABP ΔEF, AhABP ΔAS and AhABP ΔCT by transient transfection resulted in the formation of ellipsoidal aggregates commonly found at cell peripheries. In Dictyostelium, the expression of low levels of the CT fragment of the 34 kDa protein induced the formation of paracrystalline actin inclusions resembling Hirano bodies in terms of ultrastructure and composition (Maselli et al., 2002). Also, the expression of Dictyostelium 34 kDa ΔEF2 in D. discoideum and in cultured mouse fibroblasts initiated the formation of Hirano bodies, suggesting that a failure to regulate the activity or affinity of an actin cross-linking protein can provide a signal for the formation of these aggresomes (Maselli et al., 2003). It has been postulated that the formation of Hirano bodies is a general cellular response to or a consequence of an aberrant actin cytoskeletal function, which provides some insight of the delicate balance between formation and disassembly of the crosslinked actin structure necessary for the proper function of an F-actin cytoskeleton (Maselli et al., 2002). This result,suggests that EF-hands, the actinbinding site, and the carboxyl terminal contribute to the physical and structural properties of the cytoplasm.
The presence of 2 EF-hands in AhABP suggests that calcium can regulate the actin binding and other functions of this protein. AhABP is unlike most actin bundling proteins in terms of its calcium sensitivity. Our co-sedimentation assay and negative electron microscopy results show that AhABP could still bind actin in the presence of up to 10 µM of free calcium. Although AhABP has a binding activity that is similar to those of other bundling proteins in terms of its binding characteristics, it differs from these bundling proteins with respect to its response to increased calcium. Calcium is an important mediator of signal transduction pathways in a variety of eukaryotic cells, including protozoa (Scheibel, 1992) and it is critically required for the progression and completion of phagocytosis, growth, and host-parasite interactions (Burleigh and Andrews, 1998; Camacho, 2003). Furthermore, the amount of free calcium in the cytoplasm could regulate the consistency and contractility of cytoplasmic extracts and the motility of living cells, particularly in terms of producing the force required for movement. The amount of free calcium bound by AhABP conforms to the physiological level and ranges from 5-10 µM, which suggests AhABP responds to calcium levels in the normal range.
The AhABP has 2 calcium binding EF-hands, which conform with the consensus sequence for EF-hands identified by Kretsinger (Kretsinger, 1987). The replacement of glycine with isoleucine at the central position of the loop in EF-hand 1 of Dictyostelium 34 kDa actin bundling protein may caused perturb its calcium binding activity (Fechheimer et al., 1990). Interestingly, this replacement is also shown in the EF-hand 1 of AhABP, but actin binding was still exhibited under the same conditions used for Dictyostelium 34 kDa protein. The binding of calcium to a target protein often induces a conformational change of the EF-hands that triggers Ca2+-dependent cellular processes (Ikura, 1996). It is proposed that AhABP renders a semi-open conformation when it reacts with calcium. In α-actinin-titin complex, the semi-open conformation suggests a general structural solution for calcium-independent recognition by EF-hand domains. Moreover, the absence of IQ motifs in the EF-hands of AhABP, a characteristic it shares with the α-actinin-titin complex, supports that it has a semi-open calcium-independent conformation. Although, calcium-independent proteins are not regulated by calcium, they still mediate protein-protein recognition during essential cellular functions (Atkinson et al., 2001).
Many actin cross-linking proteins direct cross-linking or the bundling of actin filaments by utilizing 2 discrete actin-binding sites, thus facilitating the formation of filament bundles, branching filaments, and 3 dimensional networks (Dos Remedios et al., 2003; Puius et al., 1998). The carboxyl terminus of AhABP is suggested to function as a second actin-binding site. This is supported by weak bundling activity, the formation of thin parallel bundles, and the presence of aggresomes within cell expressing ΔCT. The fact that AhABP is a novel actin-bundling protein that can bind actin in the presence of micromolar amounts of calcium, and that it is present in cells at high concentrations suggest that it is a physiologically important component of the cytoplasmic network.
References
Notes
This work was supported by a Bio-Medical Research Institute grant, Kyungpook National University (2001).