Prevalence and Multilocus Genotyping of Giardia lamblia in Cattle in Jiangxi Province, China: Novel Assemblage E Subtypes Identified
Article information
Abstract
Giardia lamblia is a common enteric pathogen associated with diarrheal diseases. There are some reports of G. lamblia infection among different breeds of cattle in recent years worldwide. However, it is yet to know whether cattle in Jiangxi province, southeastern China is infected with G. lamblia. The objectives of the present study were to investigate the prevalence and examine the multilocus genotypes of G. lamblia in cattle in Jiangxi province. A total of 556 fecal samples were collected from 3 cattle breeds (dairy cattle, beef cattle, and buffalo) in Jiangxi province, and the prevalence and genotypes of G. lamblia were determined by the nested PCR amplification of the beta-giardin (bg) gene. A total of 52 samples (9.2%) were positive for G. lamblia. The highest prevalence of G. lamblia was detected in dairy cattle (20.0%), followed by that in beef cattle (6.4%), and meat buffalo (0.9%). Multilocus sequence typing of G. lamblia was performed based on sequences of the bg, triose phosphate isomerase and glutamate dehydrogenase loci, and 22, 42, and 52 samples were amplifiable, respectively, forming 15 MLGs. Moreover, one mixed G. lamblia infection (assemblages A and E) was found in the present study. Altogether, 6 novel assemblage E subtypes (E41*–E46*) were identified for the first time. These results not only provided baseline data for the control of G. lamblia infection in cattle in this southeastern province of China, but also enriched the molecular epidemiological data and genetic diversity of G. lamblia in cattle.
Giardia lamblia is a common zoonotic parasite, causing intestinal diseases such as diarrhea in humans and animals. In the life cycle of this parasite, cysts are spread directly through feces by infected humans or animals [1]. The transmission route is via fecal-oral or by ingesting food and water contaminated with cysts [1]. G. lamblia can cause giardiasis, which may lead to asymptomatic infection or abdominal pain, chronic diarrhea, and vomiting seriously impacting on health condition, resulting in low productivity in ruminants [2]. Cattle is one of the main hosts of G. lamblia that can excrete cysts, which is the potential threat to humans [3].
Molecular techniques have been applied for G. lamblia detection [4]. To date, 8 assemblages (A to H) of G. lamblia have been identified from different hosts. Among these assemblages, assemblages A and B are considered to be zoonotic, while other assemblages are mostly host specific [1]. In addition, assemblage E is the dominant genotype of G. lamblia occurring in cattle [5–8], and zoonotic assemblages A and B have also been identified in cattle and humans in some studies [9–13]. However, there are some studies that have found assemblage E existing in human cases [5,14,15].
Despite the widespread presence of G. lamblia in cattle, the clinical significance of this organism in cattle is not fully understood. G. lamblia infection is associated with the occurrence of diarrhea in the cattle [8,16]. Recently, a study showed that diarrhea cattle is more likely to be infected with pathogens than non-diarrhea animals [17]. Furthermore, healthy cattle are also infected with G. lamblia [7,8]. Therefore, the prevention is more important than treatment. Multilocus sequence typing has been used to identify multilocus genotypes (MLGs) of G. lamblia in dairy cattle based on sequences of the beta-giardin (bg), glutamate dehydrogenase (gdh) and triose phosphate isomerase (tpi) [18]. But most of previous studies have identified G. lamblia in cattle by a single gene locus only [19].
Jiangxi province is located on the south bank of the middle and lower reaches of the Yangtze River, with a population of more than 40 million people. It is one of the main beef producing provinces in southern China, with a large number of cattle being sold to southeast coastal cities. To date, there was no report of G. lamblia infection in cattle in Jiangxi province. Thus, the objectives of the present study were to investigate the prevalence and examine the MLGs of G. lamblia in cattle in Jiangxi Province, southeastern China.
A total of 556 fecal samples were randomly collected from cattle in 8 farms of Jiangxi province, southeastern China, including Nanchang city (2 dairy cattle farms and 1 beef cattle farm), Gao’an city (2 beef cattle farms), Xinyu city (1 meat buffalo farm) and Ji’an city (1 beef cattle farm and 1 free-range farm for beef cattle). This study was approved by the Animal Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences. All cattle were handled in strict accordance with good animal practice according to the Animal Ethics Procedures and Guidelines of the People’s Republic of China. All samples were obtained immediately after defecation by PE gloves and were stored in separate 50 ml centrifuge tubes containing 2.5% potassium dichromate, then packed into the box with ice and sent to the laboratory. Feces were stored at 4°C until extraction of genomic DNA. The information of each fecal sample was recorded, including geographical location, age, breed and collection date.
The stool samples were washed with double distilled water to wash off potassium dichromate, then 200 mg of each stool sample was packed into a 2 ml centrifuge tube for genomic DNA extraction using a commercial E.Z.N.A® Stool DNA kit (Omega Bio-Tek Inc., Norcross, Georgia, USA) according to the manufacturer’s protocols. The obtained genomic DNA samples were stored in −20°C until PCR amplification.
Giardia lamblia in fecal samples were identified by PCR-based sequencing of the bg locus [20]. All the bg-positive samples were then amplified to further identify MLGs of G. lamblia in cattle using primers for the gdh and tpi gene loci [20,21] (Supplementary Table S1). The primary PCR reaction mixture contained genomic DNA (2 μl), 10× Ex Taq Buffer (Mg2+–free) (2.5 μl), MgCl2 (2 mM), dNTP Mixture (0.2 mM), TaKaRa Ex Taq (0.625 U) (TaKaRa Shuzo Co., Ltd) and each primer (0.4 μM) in a total volume of 25 μl. The 2 μl of primary PCR product as template was amplified by the secondary PCR with the same reaction conditions. The positive (G. lamblia DNA) and negative (deionized water) control were added in each PCR amplification. The secondary PCR products were subsequently detected by electrophoresis in 1.5% (w/v) agarose gels with ethidium bromide.
All positive second-PCR products were sent to Xi’an Qingke Biotechnology Company for 2-directional sequencing. The Chromas v.2.6. was used to check the sequencing chromatograms. The obtained sequences were aligned with relevant sequences available in GenBank database (http://www.ncbi.lm.nih.gov/GenBank) and were analyzed by the Clustal X 1.81 [22] to identify the genotypes of G. lamblia. The nucleotide sequences of novel subtypes in assemblages were deposited to the National Center for Biotechnology Information GenBank database under accession numbers: MT123524 for the bg locus, MT123525–MT123526 for the gdh locus, and MT123527–MT123529 for the tpi locus. The phylogenetic relationships among G. lamblia isolates were re-constructed based on gdh and tpi sequences using a Neighbor-Joining algorithm [23] in DNAStar 5.0 [24], and the Kimura 2-parameter analysis (1,000 replicates) was selected.
The difference in G. lamblia prevalence among different breeds, regions and ages of cattle was assessed using the Chi-square (χ2) analysis by IBM SPSS Statistics for Windows, Version 22.0 (IBM Armonk Corp., Armonk, New York, USA), and P-value of 0.05 was considered as the threshold for significant difference.
In this study, G. lamblia was detected in 9.2% (95% CI 6.81–11.57) of 556 cattle fecal samples from 4 cities of Jiangxi province, with the prevalence ranging from 0.9% to 16.2% (Table 1). The highest G. lamblia prevalence (16.2%, 95% CI 11.37–21.06) was detected in cattle in Nanchang city, which was significantly higher than that in Xinyu city (0.9%, 95% CI 0–2.73) (P<0.01). The different prevalence could be due to different feeding methods. Moreover, dairy cattle (20%) and beef cattle (6.4%) were more susceptible to G. lamblia than buffalos (0.9%), for which the ORs were 26.75 (95% CI 3.60–198.79) and 7.27 (95% CI 0.96–55.13), respectively (Table 1). This result may be related to different management and sanitation status of different farms, which suggests that we should pay more attention to control G. lamblia transmission in dairy cattle and beef cattle in investigated areas. Furthermore, the highest G. lamblia prevalence was detected in cattle aged 1–3 months (19.6%, 95% CI 8.10–31.03), which is consistent with that of previous studies [25–27]. Cattle aged 1–3 months (19.6%) had a significantly higher G. lamblia prevalence compared with cattle aged more than 24 months (7.5%) (P<0.05) (Table 1). This finding illustrates that calves are more susceptible to G. lamblia infection.

Prevalence and factors associated with Giardia lamblia infection in cattle in Jiangxi Province, China
The overall G. lamblia prevalence (9.2%) in cattle in this study was higher than that in cattle in Zhaoqing city, Guangdong Province (2.2%) [28], and Qinghai Province, China (5.2%) [29], an arid area of central Iran (4.2%) [30], Thailand (6.0%) [31], and Urmia, northwest Iran (9.34%) [32]; but lower than that in Guangdong Province (74.2%) [6], Hubei Province (22.6%) [33] and Sichuan Province, China (9.4%) [34]; Egypt (13.3%) [35], South Korea (10.0%) [36], Ethiopia (9.6%) [37] and Vietnam (13.8%) [38]. The difference in G. lamblia prevalence may be related to difference in study design, breed and age of cattle studied and sampling size.
In the present study, 52 positive samples represented 2 assemblages (assemblage E and assemblage A) (Tables 2, 3), containing one novel subtype of assemblage E (named as E41*, n=5) based on sequence analyses of the bg locus (Table 2). Moreover, 42 of the 52 bg-positive samples were tested positive at the gdh locus, and 22 samples were tested tpi-positive. Two novel subtypes (named as E45*, n=4 and E46*, n=1) and 3 novel subtypes (named as E42*, n=5; E43*, n=2 and E44*, n=1) of assemblage E were identified at the gdh and tpi loci, respectively (Table 2). Previous studies have shown that the assemblage E is the primary genotype in different animals [1,39–45]. Both assemblage A and assemblage E were also found in humans [1,5]. These findings suggested that cattle could be reservoir host of G. lamblia and transport this zoonotic pathogen to humans and other animals. MLG analysis can assess the genetic diversity of G. lamblia [46]. Among these 52 samples, 16 samples were successfully amplified and sequenced at all the 3 genetic loci (bg, gdh, and tpi) (Table 4), forming 15 novel MLGs (named as MLGE1-E14, MLGA1) and one mixed infection (Table 4), which revealed a high genetic diversity of G. lamblia assemblages.
The phylogenetic tree based on G. lamblia tpi-sequences and gdh-sequences were constructed in order to evaluate the genetic relationships of the G. lamblia isolates. The results showed that G. lamblia isolates from cattle (novel subtypes E41*–46*) were clustered within the assemblage E (Figs. 1, 2), which contained G. lamblia isolates from animals and humans with a higher confidence value, revealing closer genetic relationship (Fig. 1).
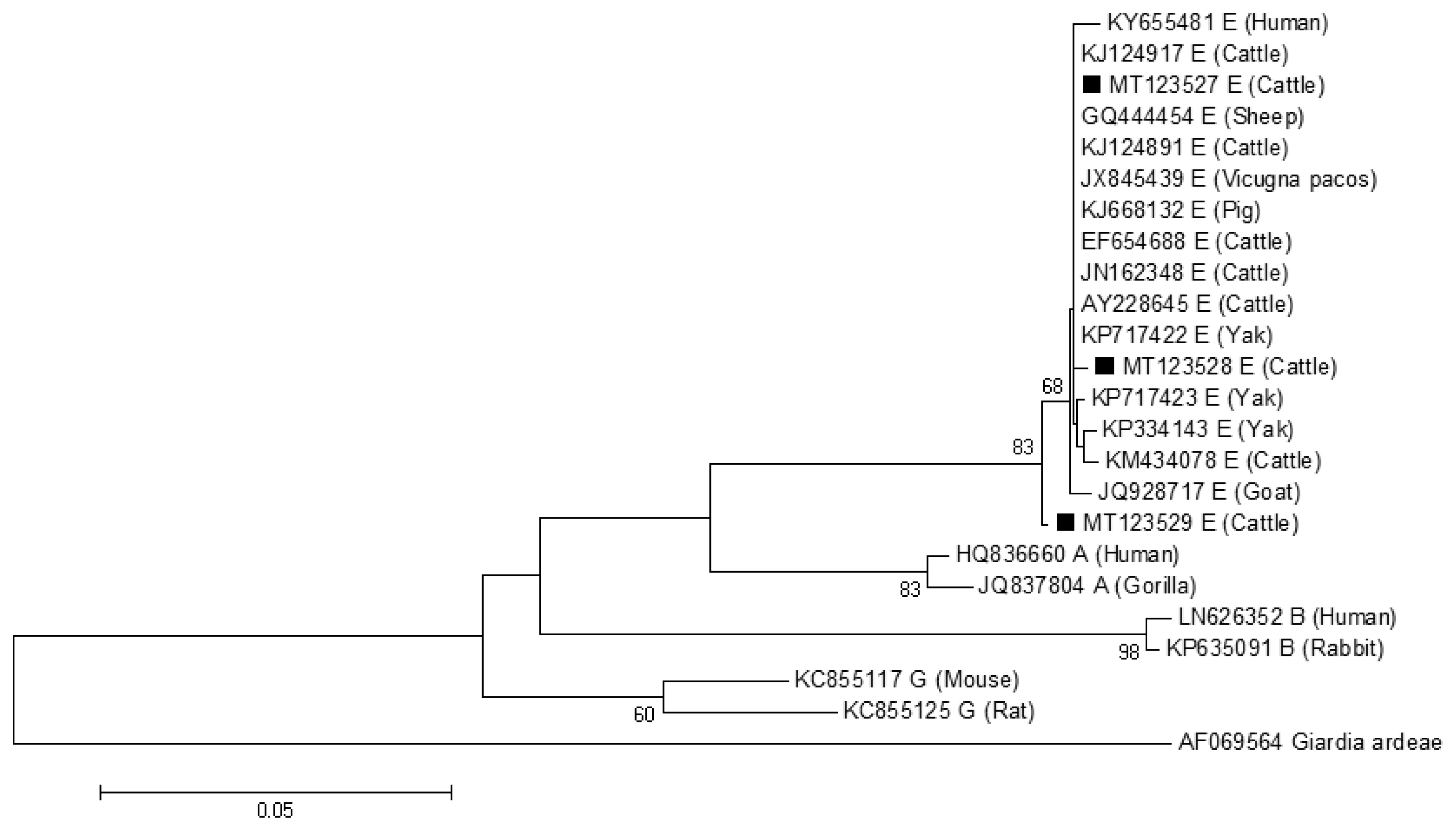
A phylogenetic tree of Giardia lamblia tpi gene sequences. Novel subtypes of assemblage E isolates identified from cattle in this study are marked with a black square (■).
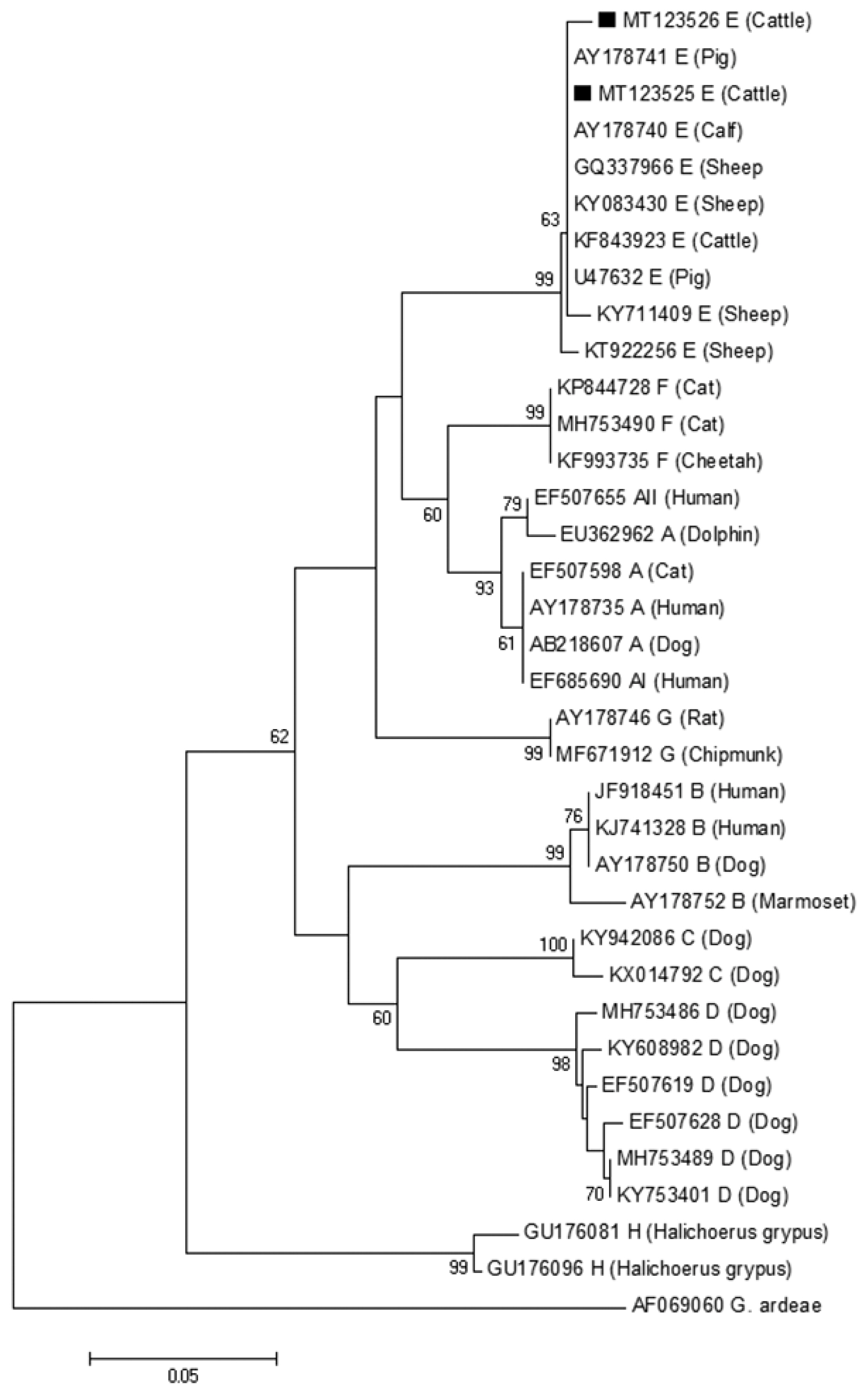
A phylogenetic tree of Giardia lamblia on gdh gene sequences. The isolates marked with a black square (■) represent novel subtypes of assemblage identified from cattle in this study.
In conclusion, this study revealed a 9.2% G. lamblia prevalence in cattle in Jiangxi province, southeastern China, and identified 6 novel assemblage E subtypes (designated as E41*–46*) for the first time. Fourteen novel MLGs within assemblage E and one novel MLG within assemblage A were identified. These findings revealed the prevalence and MLGs of G. lamblia in cattle in Jiangxi province, which not only enriched the genetic diversity of G. lamblia, but also provided baseline data for controlling G. lamblia infection in animals and humans in this southeastern province of China.
Supplementary Information
Nested PCR primers, annealing temperature, and expected fragment size for the detection of Giardia lamblia in cattle
ACKNOWLEDGMENT
Project support was provided by the Agricultural Science and Technology Innovation Program (ASTIP) (Grant No. CAAS-ASTIP-2016-LVRI-03) and the Elite Program of Chinese Academy of Agricultural Sciences.
Notes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.



