Genetic diversity of Plasmodium falciparum erythrocyte membrane protein 1 in field isolates from central Myanmar
Article information
Abstract
Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1), encoded by the polymorphic var multigene family, is a highly polymorphic antigen that plays a crucial role in the pathology of malaria. The contribution of the genetic diversity of var toward the immune escape of P. falciparum has not yet been fully elucidated. This study aimed to characterize the diversity of var repertoires by screening P. falciparum Duffy-binding-like α domain (PfDBLα) among field isolates from central Myanmar. Genetic analysis revealed that the D-H segments of var in Myanmar populations have an extensive polymorphic repertoire, with high numbers of unique sequence types in each individual. However, var genes from the global population, including Myanmar, shared close genetic lineages regardless of their geographic origins, indicating that they have not undergone rapid evolutionary changes.
Introduction
Malaria, one of the most serious vector-borne infections, is caused by Plasmodium parasites. In 2020, 241 million people were diagnosed with malaria, of which 627,000 died. Half of the world’s population is at risk of contracting the disease [1]. Malaria parasites exploit the diversity of their surface antigens to evade the host’s immune system, which is a major impediment to the development of effective malaria vaccines [2].
Several Plasmodium species contain multigene families encoding variant surface antigens involved in cell binding. P. falciparum erythrocyte membrane protein 1 (PfEMP1), encoded by the polymorphic var multigene family, is a well-characterized variant antigen expressed on the surface of infected erythrocytes and implicated in the pathology of malaria [3]. Although PfEMP1 is highly variable among parasite isolates, only one type is expressed on the surface of infected red blood cells. It is either stably inherited through successive cell cycles or switched off by the expression of a different gene during infection [4,5].
PfEMP1 comprises domain assemblies that vary in their composition and sizes and is arranged in intra- and extracellular regions. Each member of the multicopy var family contains 2 exons (Fig. 1). Exon 2, approximately 1.5-kb long, is highly conserved and encodes a putative intracellular region called the acidic terminal segment, whereas exon 1, approximately 4–10-kb long, is highly variable and encodes a putative extracellular region and a transmembrane domain [6]. The extracellular region predominantly comprises an N-terminal segment, a duplicated arrangement of Duffy-binding-like (DBL) domains, tandem cysteine-rich interdomain regions (CIDRs), and C2 domains. The DBL and CIDR domains form 5 (α, β, γ, δ, and ɛ) and 3 (α, β, and γ) types, respectively, in which sequences have unique specific consensus motifs that can be used to characterize distinct DBL-CIDR domains. The prototypical head structure of most PfEMP1 members includes a DBL1α-CIDR1α semi-conserved tandem, followed by DBLδ-CIDRβ [6–10].
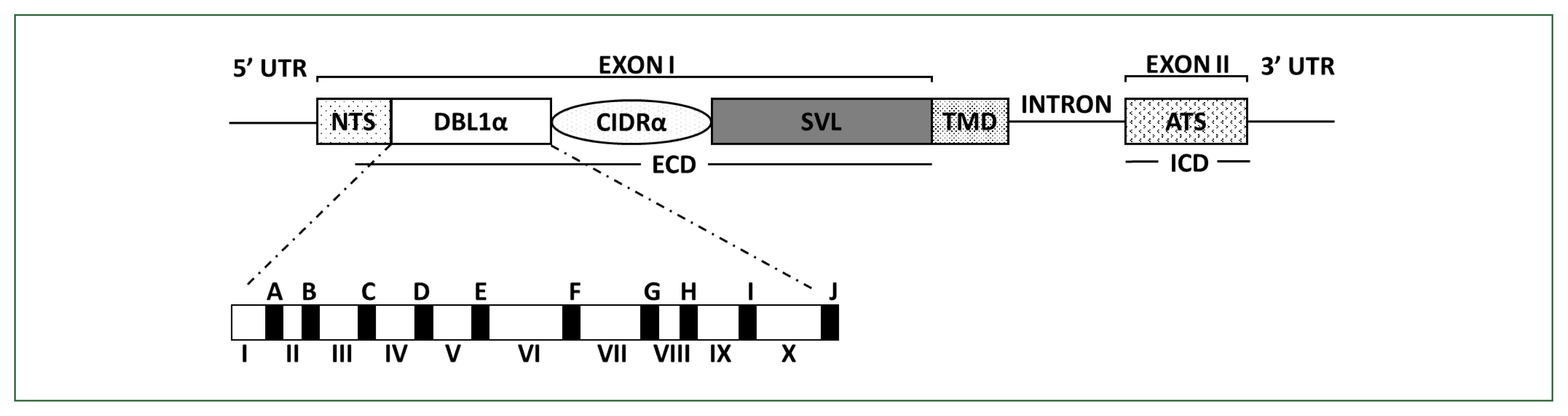
Schematic overview of PfEMP1 and DBLα domain structures encoded by var. PfEMP1 comprises an extracellular domain (ECD), a transmembrane domain (TMD), and an intracellular acidic terminal segment (ATS). The ECD varies the most in its composition and size. In the DBL1α domain, conserved sequences are labeled from A to J (filled blocks), and hypervariable sequences are labeled from I to X (unfilled blocks), respectively. PfEMP1, Plasmodium falciparum erythrocyte membrane protein 1; CIDR, cysteine-rich interdomain region; DBL, Duffy-binding-like domain; NTS, N-terminal segment; SVL, segment of variable length.
Malaria distribution in Myanmar is heterogeneous and is influenced by several factors, such as the environment, population geospatial repartition, and international and internal migration [11–13]. Some molecular epidemiological studies have been conducted on P. falciparum isolates collected from the border areas of Myanmar [14,15]; however, information on the genetic diversity of PfEMP1 among isolates from other regions of Myanmar is lacking.
This study aimed to characterize the genetic diversity of var repertoires. We screened PfDBLα in P. falciparum isolates collected from central Myanmar. Understanding the nationwide diversity of PfEMP1 can provide insights into the genetic structure of the parasite and the root of persistent disease transmission, ultimately aiding in the successful elimination of the parasite.
Materials and Methods
Ethics statement
This study was approved by the Ethics Committee of the Ministry of Health, Myanmar (97/Ethics 2015) and Kyungpook National University (2017–0010). Written informed consent was obtained from each participant. For individuals aged <18 years, consent was obtained from their parents.
Study sites and sample collection
Samples were collected from the central areas of Myanmar, including Mandalay, Naung Cho, Pyin Oo Lwin, and Tha Beik Kyin, from patients diagnosed with P. falciparum infections between August 2013 and December 2015. Falciparum malaria was diagnosed using a rapid diagnostic test and a subsequent microscopic examination followed by polymerase chain reaction (PCR) for confirmation [16]. A total of 48 blood samples from P. falciparum-infected patients (42 men and 6 women, age 3–50 years) were analyzed in this study (Supplementary Table S1). Genomic DNA (gDNA) was purified from the samples using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany).
PCR amplification of PfDBLα
PfDBLα was amplified from the gDNA of each isolate using previously used degenerate primer sets BF (forward: 5′-GCMTGYGCDCCRTWYMGAMG-3′) and BH (reverse: 5′-CKGCCCATTCYTCRAACCA-3′) targeting the semi-conserved blocks B and H of DBLα [3,17]. Briefly, PCR was performed with the following conditions: 94°C for 2 min (initial denaturation); 35 cycles at 94°C for 5 sec, 50°C for 20 sec, and 60°C for 45 sec; and 60°C for 2 min (final extension) [18]. The PCR results were confirmed by electrophoresis on a 1.5% agarose gel, followed by staining with ethidium bromide and visualization under ultraviolet light.
Cloning and sequencing
For each isolate, amplicons scattered in the 450–700-bp region were gel-purified, ligated into the pGEM-T Easy Cloning Vector Kit (Promega, Madison, WI, USA), and transformed into Escherichia coli DH5α competent cells according to the manufacturer’s instructions. Twenty to forty colonies per each cloned amplicon were selected and sequenced using Sanger’s method (Macrogen Inc., Daejeon, Korea). The obtained sequences were aligned with global sequences from GenBank and analyzed using CLC Main Workbench 6.
Sequence analysis
Raw sequence data were obtained by excluding all nonspecific and low-quality sequences as well as human or vector contamination of each clone sequence. The PfEMP1-encoding sequences were identified from the high-quality sequences using BLASTx (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastx&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome). Clones acquired from the same isolate and with a DBLα nucleotide sequence identity of at least 96.0% were classified into the same sequence type (ST) of var. DNA sequences were translated using EMBL-EBI (https://www.ebi.ac.uk/Tools/st/emboss_transeq/), and amino acid sequences were aligned against those hosted on the 3D7 genome database of the PlasmoDB interface (https://plasmodb.org/plasmo/) using BLAST to categorize their homology groups.
Var sequence data from the global population
The global var sequence data used for the comparison study were downloaded from GenBank and corresponded to 3 regions: Southeast Asia, Sub-Saharan Africa, and South America. In total, 73 sequences were obtained from Papua New Guinea, Thailand, and Indonesia (Asian population); Gabon, Kenya, Mali, Malawi, Sudan, Tanzania, and Senegal (African population); and Brazil, Venezuela, and Honduras (South American population). Additionally, 38 var sequences from the 3D7 strain were obtained from PlasmoDB. Accession numbers for the downloaded sequences are listed in Supplementary Table S2.
Phylogenetic analysis
The phylogenetic tree was constructed using MEGA X (https://www.megasoftware.net/) with the minimum-evolution method and the maximum composite likelihood model. Evolutionary analyses were performed on the Myanmar var sequences using the 3D7 strain and global populations as references.
Results
PfDBLα sequence types
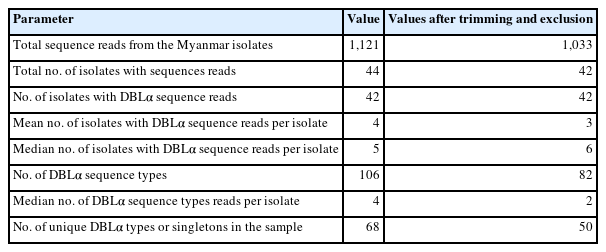
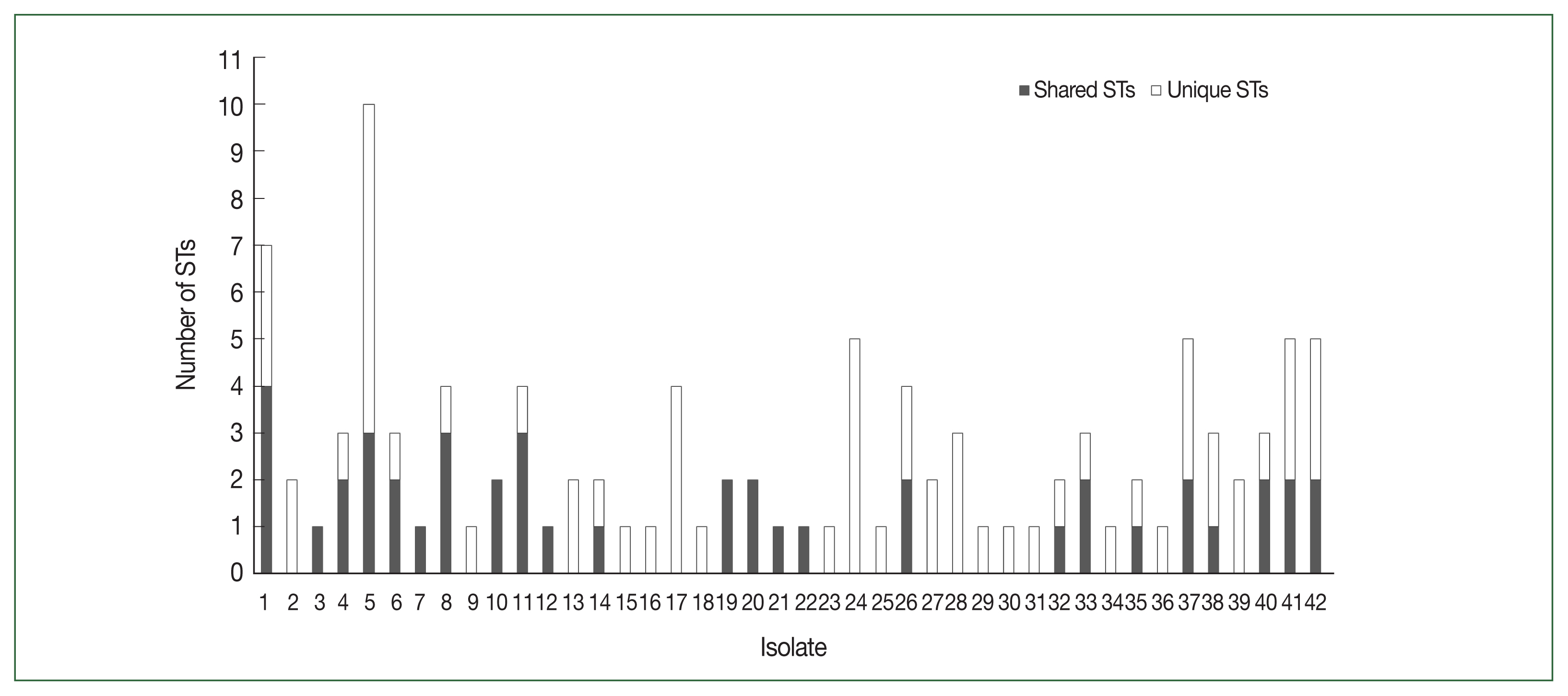
Of the 48 Myanmar P. falciparum isolates, 42 successfully yielded DBLα reads. After applying the exclusion criteria, 1033 individual BF/BH inserts were successfully sequenced, resulting in 82 distinct DBL STs (Table 1). The PfEMP1-encoding sequences obtained have been deposited in GenBank (Accession Nos. ON991795–ON991950). Most isolates had multiple STs with unique sequences that occurred only once in an isolate (singletons). The number of distinct DBL var sequences detected per isolate varied from 1 to 20. The distribution of STs in the study population is shown in Fig. 2.
Distribution and frequency of PfDBLα groups
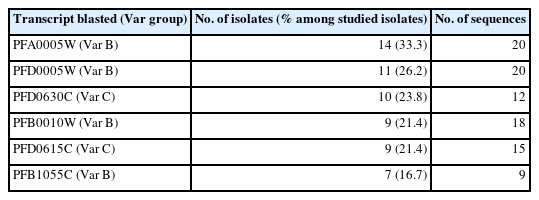
The DBLα sequence from each isolate was aligned against the sequences hosted on the 3D7 genome, and the sequence with the highest alignment score was assigned the name of the gene as identified from the 3D7 genome (Table 2). Thirty-nine different 3D7 homologs containing PfDBLα-encoding sequences were identified. Transcripts from the DBLα groups A, B, C, B/A, and B/C were detected in isolates, and those from groups B and C were mostly shared. The highly dominant DBLα sequences were confirmed to belong to var B, including the transcripts PFA0005W (33.3%), PFD0005W (26.2%), and PFB0010W (21.4%), followed by var C, including the transcripts PFD0630C (23.8%) and PFD0615C (21.4%; Table 2).
Genetic diversity and phylogenetic analysis of PfDBLα in Myanmar and global populations
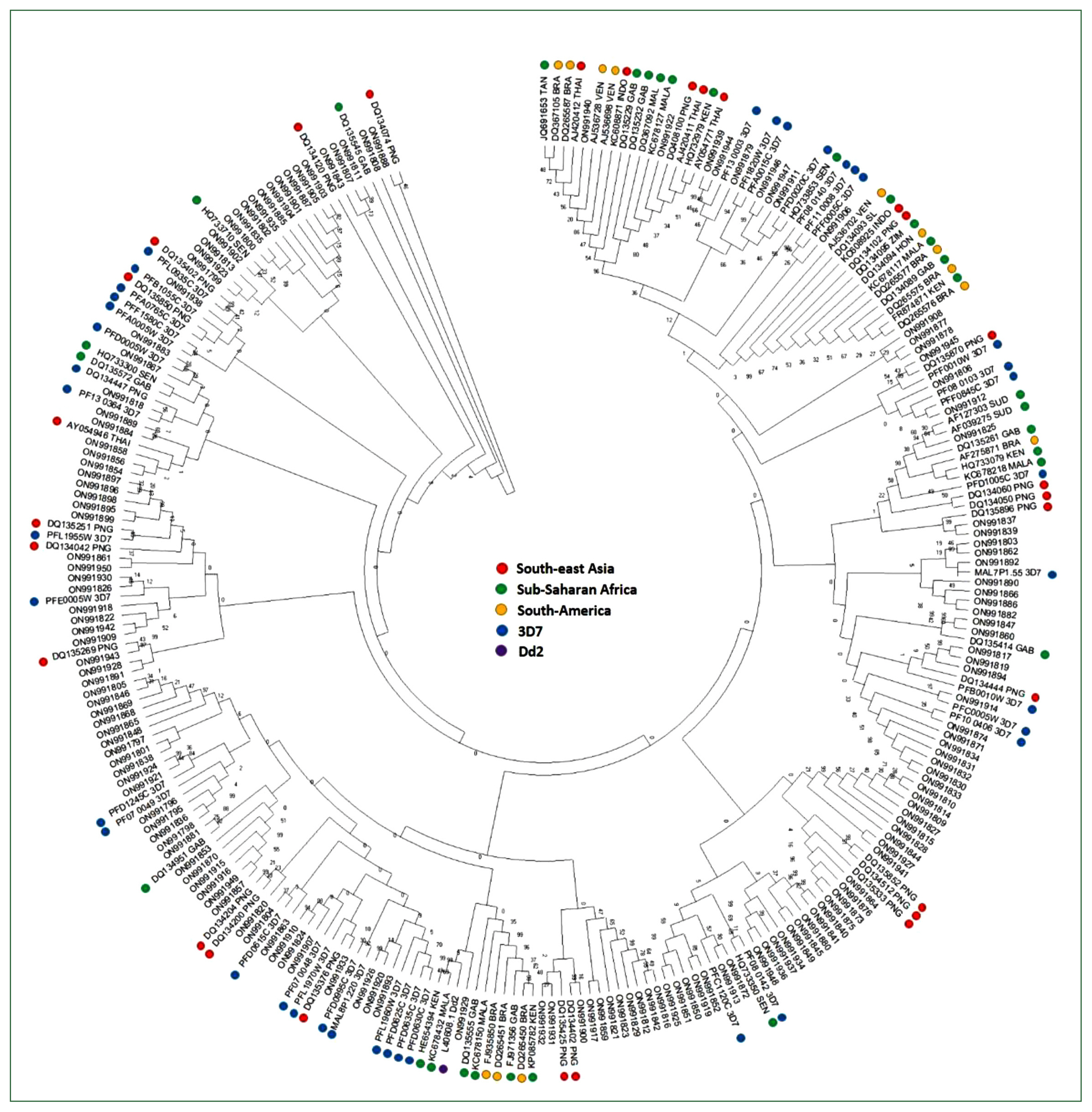
The genetic diversity of the studied population was compared by aligning against the D-H segment of the P. falciparum 3D7 var genes. All individuals with DBLα exhibited conserved cysteine-rich motifs at all positions (Supplementary Fig. S1). Var sequences from the Myanmar population were highly diverse and clustered with the global var genes used as a reference by sharing the bootstrap root from the Asian, European, African, and American var sequences (Fig. 3). In addition, the D-H segment sequences of var genes from all continents (Asia, Europe, Africa, and America) and the 3D7 strain were distributed in all branches without any region-specific cluster.
Discussion
Myanmar bears the high burden of malaria in the Greater Mekong Subregion. Although the recent incidence of malaria in the country has decreased, the genetic diversity of malaria parasites remains high, making it an important public health problem. Asymptomatic infections and the spread of antimalarial drug resistance also are major concerns [13,15]. In addition, the level of malaria transmission in Myanmar varies by region, making the regional persistence of the disease heterogeneous [19–22]. Therefore, understanding the genetic diversity of malaria parasites represents a major step toward disease eradication because it can provide insights into effective vaccine development.
In this study, we investigated the genetic diversity of var genes within a natural P. falciparum population in central Myanmar by analyzing their DBLα domain-encoding sequences. This particular sequence motif was chosen because DBLα domains are used as polymorphic markers in gene diversity and gene expression studies. Moreover, hyperendemic regions, such as DBLα domains, directly help the parasite evade the immune response [17,23, 24]. A high degree of var recombination increases the genetic diversity between isolates in neighboring and distant geographic locations [6,25]. Therefore, examining the genetic diversity of DBLα-encoding sequences can provide valuable information about the molecular structure of the P. falciparum population in Myanmar. An extensive range of var repertoires was found among the studied population. After applying a 96.0% cutoff to define the DBLα types with a minimum sequence overlap, 106 STs were identified, and singletons occurred at a high frequency in the population (Fig. 3). The mean number of shared ST sequences among isolates was not significant, suggesting a constant event in the var recombination repertoire and extensive diversity.
The var-encoded hypervariable PfEMP1 family of proteins mediates the adhesion of infected erythrocytes to various host cells during the blood stage of malaria infection. Most PfEMP1-encoding var genes are located in the subtelomeric regions, whereas others are located centrally in chromosomes [26]. Based on their sequence similarity and analysis of the intron and 5′ and 3′ untranslated regions, var genes can be categorized into 3 major subgroups (var groups A, B, and C) and 2 intermediate groups (B/A and B/C), which represent transitions between the 3 main groups [27]. Cham et al. reported that children progressively acquired a broader repertoire of anti-PfEMP1 antibodies in the background of high exposure to malaria infection, but as their age increased, the identified antibodies acquired against particular DBL-like domains belonged to groups other than A or B/A [28]. Therefore, in individuals frequently exposed to malaria, parasites express higher levels of groups B, C, or B/C PfEMP1, which correlates with their reduced fitness and pathogenicity during subsequent infections [28]. The population examined in this study was within the working age (93.0%), and var group B predominated, followed by var group C (Table 2). The expression of a restricted and antigenically semi-conserved subset of PfEMP1 suggests that this population has acquired a minimum level of immunity against malaria infection because of being constantly exposed to the disease.
In this study, within individual patients, several DBL α sequences were found more frequently than others but they were also shared between individuals. The DBLα domain contains 16–18 conserved cysteine residues, which likely play an important role in its folding and structure [29]. The number of cysteine residues determines the classification of the DBLα domain and the severity of the disease [6,30]. Polymorphism within the hypervariable region induced via gene recombination and/or duplication results in length and sequence variability among parasites between and within hosts in malaria-endemic areas [31]. The DBLα domain has an average of 4 cysteine residues, and the number can vary from 2 to 9. Variants with unusual numbers of residues may exhibit altered antigenic and adhesive properties. Among the Myanmar population studied, the cysteine at the CRC motif was almost conserved in all the var genes and was highly conserved in the homology blocks VWKAiTC and FRkTC. The semi-conservation of cysteine residues within the Myanmar sequences suggests that cysteine contributes toward parasite maintenance in these low-endemic areas. Even with the low number of patients included in this study, the targeted DBL1α domain showed a global distribution of var within the central regions of Myanmar. On account of sharing borders with China, India, and Thailand, we expect to find predominant geographical var types related to any or all of these countries. However, these results highlight the constancy of population movement in and out of different countries that share borders with Myanmar [32]. Consistent with the conserved cysteine residues among populations, phylogenetic analysis of var genes from Myanmar and global isolates revealed that the D-H segment was not distributed in region-specific clusters, although the gene showed genetic diversity within isolates.
Supplementary Information
Characteristics of the studied population
List of sequence data used for phylogenetic analysis
Alignment of the DBLα sequences of Myanmar population and 3D7 strain.
Acknowledgment
This research was financially supported by the Basic Science Research Program (NRF-2019 R1C1C1002170) and Brain Pool program (NRF-2018H1D3A1A02074759) funded by the Ministry of Science, ICT and Future Planning through the National Research Foundation of Korea (NRF).
Notes
The authors declare no conflict of interest related to this study.
Author contributions
Conceptualization: Chung DI, Hong YC, Na BK, Goo YK
Data curation: Dinzouna-Boutamba SD, Lee S, Goo YK
Formal analysis: Dinzouna-Boutamba SD, Lee S, Moon Z
Funding acquisition: Goo YK
Investigation: Dinzouna-Boutamba SD, Lee S, Na BK
Resources: Chung DI, Myint MK, Naw H, Na BK
Validation: Naw H,
Visualization: Lee S
Writing – original draft: Dinzouna-Boutamba SD, Lee S, Goo YK
Writing – review & editing: Na BK, Goo YK