Detection of intestinal parasites in leopard cat fecal samples using shotgun metagenomics
Article information
Abstract
The leopard cat (Prionailurus bengalensis) is a wild felid species that serves as a reservoir of zoonotic parasites. In this study, we investigated intestinal parasite taxa by reanalyzing previously published shotgun metagenomic sequencing data from fecal samples of wild leopard cats using a custom 18S rRNA gene reference database constructed from the NCBI nucleotide database. Among 11 metagenomic samples, 5 parasite species were identified: Toxoplasma gondii, Clonorchis sinensis, Strongyloides planiceps, Cylicospirura petrowi, and Pharyngostomum cordatum. These findings demonstrate that shotgun metagenomic analysis of fecal samples can be a useful tool for monitoring zoonotic parasite infections in this species and for investigating parasite life cycles. However, this approach is limited by its dependence on existing reference databases and requires experimental validation of the findings.
The leopard cat (Prionailurus bengalensis) is one of the most widely distributed wild felids in Korea. As a free-ranging carnivore that preys on a variety of small mammals, birds, amphibians, and reptiles, the leopard cat plays an important role in maintaining ecological balance. However, it also serves as a potential reservoir for zoonotic pathogens that can influence public health, including intestinal parasites. In Korea, previous studies identified diverse parasitic infections in leopard cats, including by helminths such as Thelazia callipaeda and Clonorchis sinensis and protozoans such as Toxoplasma gondii [1–3]. These parasites not only affect the health of wildlife but can also pose spillover risks to domestic animals and humans. Thus, the surveillance of parasitic infections in wild carnivores such as leopard cats is crucial from a One Health perspective. In this study, we identified intestinal parasite taxa by reanalyzing previously published fecal metagenomic datasets from wild leopard cats. Our findings provide updated information on the zoonotic parasite fauna associated with this important wildlife species.
Shotgun metagenomic sequencing data from 11 fecal samples of wild leopard cats collected in Chungcheongnam-do and Gyeongsangbuk-do, Korea, were downloaded from the NCBI Sequence Read Archive (BioProject accession No. PRJNA772888) [4,5]. These data were originally generated for bacterial and dietary profiling in leopard cats [4,5]. In this study, we reanalyzed metagenomic datasets for eukaryotic taxonomic profiling, focusing on the identification of intestinal parasites. All sequences annotated as “18S rRNA” in the NCBI nucleotide database (https://www.ncbi.nlm.nih.gov/nuccore/) were downloaded and used to construct the custom reference database. For database construction, we first selected complete or near-complete 18S rRNA sequences (>95% gene coverage). Second, their taxonomic assignments were verified against established parasite databases. Third, redundant sequences were removed using CD-HIT (v4.8.1) with a similarity threshold of 95% (https://github.com/weizhongli/cdhit). Final curation was performed to ensure proper representation of known parasitic taxa, consistent with our previous studies [6,7]. Metagenomic shotgun sequencing reads were processed using the QIIME 2 platform, following the official MOSHPIT user documentation (https://github.com/bokulich-lab/moshpit-docs). Reads shorter than 150 bp or with a quality score below Q30 were discarded (Supplementary Table S1). The 150 bp minimum length threshold was selected to ensure sufficient sequence information for reliable taxonomic classification of 18S rRNA sequences, as shorter fragments may lead to ambiguous assignments. This conservative filtering strategy was intended to enhance the specificity of parasite detection and minimize the risk of false-positive results.
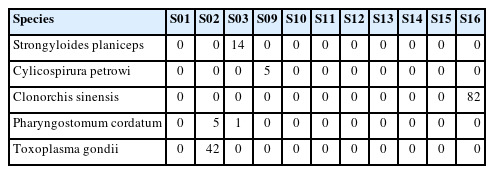
Our reanalysis of the fecal samples of 11 leopard cats identified 5 parasite taxa: Strongyloides planiceps, Cylicospirura petrowi, C. sinensis, Pharyngostomum cordatum, and T. gondii (Table 1). All reads assigned to these taxa were 151 bp in length and showed 100% query coverage and 100% sequence identity with reference sequences in the database (Table 2).
C. sinensis and T. gondii were detected in samples S16 and S02, respectively. These findings agree with previous reports highlighting the role of leopard cats as reservoirs of zoonotic parasites in Korea [1,2,6]. These parasites are also among the most commonly reported causes of human parasitic infections in the Korean population [8,9]. Infections with C. sinensis and T. gondii have been reported in stray cats in Korea, raising concerns about possible zoonotic parasite spillover among wildlife, cats, and humans [10,11].
A previous study by Woo et al. [4], who analyzed the same fecal samples from leopard cats as used in our metagenomic study, reported that the leopard cat diet includes various species of rodents and other mammals, frogs, and multiple species of fish. Fish are known as intermediate hosts for C. sinensis infection [12], while rodents and other mammals serve as intermediate hosts for T. gondii infection [13].
The low read counts (1–14 reads) for the detection of several parasites in our study warrant careful consideration of potential contamination or sequencing artifacts. However, several factors support the authenticity of these findings. All assigned reads exhibited 100% sequence identity and complete query coverage with reference sequences. Second, our conservative filtering approach, applying a Q30 quality threshold and a minimum length of 150 bp, minimizes the likelihood of erroneous assignments. Third, the identified parasite species aligned with known host ranges and geographic distribution patterns reported previously [1,2,14–17]. Nevertheless, we acknowledge that low read counts necessitate cautious interpretation, and ideally, should be validated through complementary diagnostic methods such as species-specific PCR or microscopic examination.
Non-zoonotic parasites such as S. planiceps, C. petrowi, and P. cordatum were also detected in leopard cat fecal metagenome samples. S. planiceps has previously been identified in Japan in wild carnivores such as raccoon dogs and weasels, as well as in stray cats [18]. Feline strongyloidiasis is relatively prevalent worldwide, particularly in wild felids, with a pooled prevalence of 20% [19]. Meanwhile, the gastrointestinal parasitic nematodes Cylicospirura spp. have not been reported in felids from Korea, Japan, or China, but have been reported in wild felids across Europe, North and South America, Africa, Australia, and India [20]. To the best of our knowledge, the detection of C. petrowi in this study represents the first record in Korean leopard cats and suggests that this parasite is more broadly distributed than previously thought. As a gastric parasite, C. petrowi can cause significant gastric nodules and potentially impact nutritional and immunological health in wild felids, affecting their health and survival in natural habitats. P. cordatum (Trematoda: Alariidae) has previously been reported in leopard cats from Japan and Vietnam, but in Korea it has only been detected in stray cats and in the snake Rhabdophis tigrina [14–17]. The life cycle of P. cordatum typically involves freshwater snails as the first intermediate host and amphibians or reptiles as the second intermediate host, reflecting the feeding ecology of leopard cats. The detection of this parasite provides insights into leopard cats’ predatory behaviors and their integration within local food webs.
Shotgun metagenomic analysis of stool samples offers significant advantages in detecting and identifying multiple parasitic taxa simultaneously and investigating their interactions with other host and microbial genes, but it also has limitations. This approach offers several advantages over conventional diagnostic methods. First, it provides the ability to simultaneously detect multiple parasites without the need for parasite-specific primers or specialized morphological expertise. Second, it has the potential to detect unexpected or novel parasite associations that might be overlooked in targeted studies. Third, it provides an opportunity for the retrospective analysis of existing datasets to address new research questions without additional sampling. Finally, it allows the integration of parasite data with broader microbiome analysis to understand potential interactions [4]. In particular, there is still a need to evaluate and compare the sensitivity and specificity of metagenomic approaches relative to conventional parasite diagnostic methods, such as microscopy and PCR. Moreover, only parasite species with DNA sequences already registered in the NCBI gene database could be detected, limiting the identification of unregistered or novel taxa. The generalizability of our findings is also limited by the small sample size (n=11) of the dataset and the fact that parasites were detected in only 4 samples (S02, S03, S09, and S16). Furthermore, the absence of metadata for the fecal samples, such as host age, sex, and collection season, prevents a more detailed epidemiological interpretation of the results. However, this proof-of-concept study confirms the validity of using existing metagenomic datasets for parasite surveillance. Future studies should aim for larger-scale analyses with datasets designed expressly for parasite discovery.
Notes
Author contributions
Conceptualization: Cho YH
Data curation: Choi DY, Chavarria X
Formal analysis: Yi M, Shatta A
Project administration: Oh S
Supervision: Choe S
Validation: Kang D, Lee SH
Writing – original draft: Choi JH
Writing – review & editing: Kim JY
Conflict of interest
The authors declare no conflict of interest related to this study.
Acknowledgments
This study was supported by the National Research Foundation of Korea (NRF), funded by the Korea government (MSIT) (grant No. RS-2024-00456300), and by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant No. RS-2024-00406488 and RS-2023-KH139971).
We thank the original research teams for generating and publicly sharing the metagenomic sequencing data used in this study. The relevant study has been appropriately cited in the References section [4].
Supplementary Information
Supplementary material is available with this article at https://doi.org/10.3347/PHD.25032.