Abstract
To determine the molecular phylogenic location of Plagiorchis muris, 28S D1 ribosomal DNA (rDNA) and mitochondrial cytochrome C oxidase subunit I (mtCOI) were sequenced and compared with other trematodes in the family Plagiorchiidae. The 28S D1 tree of P. muris was found to be closely related to those of P. elegans and other Plagiorchis species. And, the mtCOI tree also showed that P. muris is in a separate clade with genus Glypthelmins. These results support a phylogenic relationship between members of the Plagiorchiidae, as suggested by morphologic features.
-
Key words: trematoda, ribosomal DNA, mitochondrial DNA, phylogeny, classification
Although
Plagiorchis muris was first recovered from
Rattus norvegicus (
Seo et al., 1981) in Korea, eleven natural cases of human infection by the genus
Plagiorchis have been reported (
Chai, 1991). Molecular approaches to this worm are rare since it is difficult to acquire sufficient metacercariae. Since
P. muris is unique species of which infection in human intestine has been reported among the family Plagiorchiidae, the phylogenic relationship with other worms in Plagiorchiidae is interesting. In the present study we determined molecular phylogenic location of
P. muris with respect to other trematodes in the family Plagiorchiidae using the 28S D1 rDNA and mtCOI gene regions. It will be able to tell the consistency between morphologic and molecular phylogenic location of
P. muris in the genus
Plagiorchis.
Metacercariae of
P. muris were obtained from dragonflies and adults were recovered from infected mouse intestines (
Hong et al., 1999). Worms were kept in ethanol at -70℃ until assayed. Frozen worms were lyophilized and lyzed with lysis buffer containing 1% SDS, proteinase K (500 g/ml), and RNase at 37℃ for 2-3 hrs. DNA was extracted using the phenol/chloroform method and precipitated in ethanol. The 28S D1 rDNA and mtCOI regions were amplified by PCR using the primer sets described by Qu et al. (
1988), and by Garey and Wolstenholme (
1989). PCR was conducted using a mixed solution (25l) of the above extracted DNA (2.5l) as a template (1-50 ng), and primers (1.0l, each 10 pmole) for each genes with ~1.5 units of
Taq enzyme (TAKARA Shuzo Co., LTD, Japan) in a GeneAmp PCR System 9600 (Perkin Elmer, U.S.A.). PCR amplification was conducted over 40 cycles denaturing at 95℃ for 20 seconds, annealing at 55℃ (28S D1 rDNA) or 48℃ (mtCOI) for 30 seconds, extending at 72℃ for 30 seconds), followed by a final extension of 7 minutes.
Amplified PCR products were extracted and purified using a gel extraction kit and a PCR purification kit (QIAGEN Co., Germany) and were cloned into a pT7Blue Perfectly Blunt cloning vector using T4 DNA ligase and transformed into E.coli Nova Blue competent cells, according to the supplier's protocol (Novagen Co. USA). Positive recombinant clones were picked, and grown in 2 ml of LB broth (in the presence of 50 g/ml ampicillin) overnight at 37℃. The recombinant plasmid was screened using isopropyl—thiogalactoside (IPTG) and 5-bromo-4 chloro-3-indolyl—D-galactoside (X-gal). Positive plasmid DNA was purified using a QIAprep spin plasmid kit (QIAGEN Co., Germany). DNA sequencing was performed by the dideoxy chain termination method. Cycle sequencing reactions were performed using Thermo Sequenase dye terminator sequencing pre-mix kits (Amersham Life Science Co.). PCR products were run on an ABI 373A automated DNA sequencer (Applied Biosystems model 373A, Perkin Elmer) according to the manufacturer's instructions. The each gene was sequenced in both orientations using the universal sequencing primers T7 and U19. At least two clones were sequenced per gene; additional clones were sequenced as necessary to resolve ambiguous sites.
The analyses of 28S D1 rDNA and mtCOI region sequences were determined by comparison with those of a range of other related nematodes of the family Plagiorchiidae. NCBI (National Center for Biotechnology Information, NIH, Bethesda, USA) databases were used for sequence homology analysis using BLAST2. Multiple sequence alignments were performed using CLUSTA W program (European Bioinformatics Institute,
http://www.ebi.ac.uk/clustalw/), and the fractional GC contents of nucleic acid sequences were determined using the EMBOSS GEECEE program provided by Dr. Richard Bruskiewich (Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge, CB10 1SA, UK)(
http://analysis.molbiol.ox.ac.uk/pise_html/geecee.html). Alignment gaps were treated as missing data. Other trematodes in the family Plagiorchiidae and their GenBank accession numbers used for comparative purposes in this study are listed in
Tables 1 and
2.
Haematoloechus longiplexus (Haematoloechidae, AY222280 for 28S rDNA) and
Paragonimus macrorchis (Paragonimidae, AF159598 for mtCOI) were used as outgroups. Phylogenic analysis was performed using the Kimura 2-parameter model for distance correction; corrected for multiple substitutions (
Kimura, 1980). Phylogenic trees were constructed using the Neighbor-joining algorithm (
Swofford et al., 1996). In order to view these trees, we used PHYLIP version 1.6 for tree drawing using the parsimony, maximum likelihood, and distance methods.
The sizes of amplified genomic DNA fragments of 28S D1 rDNA and mtCOI were 0.3 kb and 0.45 kb, respectively. The sequences length was 309 bp for the 28S D1 rDNA gene, and 397 bp for the mtCOI gene excluding primer sequences (results not shown). The GC contents were 52% (28S D1 rDNA) and 48% (mtCOI) exlcuding primers. The 28S D1 rDNA and mtCOI coding regions were highly conserved by multiple sequence alignment without additional nucleotides, Overall nucleotide similarity between
P. muris and other
Plagiorchiidae species ranged 61.1%~63.7% for 28S D1 rDNA and 51.4%~63.9% for mtCOI (results not shown). Several insertions, a 79 bp insertion in
P. muris 28S D1 rDNA, a 16 bp insertion in
P. muris mtCOI, with gaps, 1 bp gap in
P. muris 28S D1, in the same or in different positions were detected within and between species (results not shown). The most-parsimonious tree was obtained when gaps were treated as missing data. The 28S D1 tree of
P. muris was closely related with that of
P. elegans and other
Plagiorchis species (
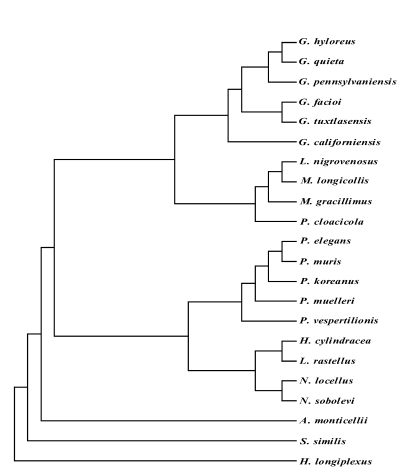
Fig. 1). The mtCOI tree of
P. muris was in the separate clade with genus
Glypthelmins (
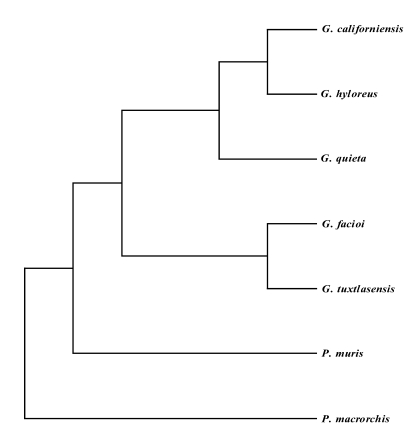
Fig. 2).
We found sequence variability in both the 28S rDNA and mtCOI region of the family Plagiorchiidae, but the 28S D1 rDNA region was more conserved than the mtCOI region. With respect to the 28S D1 rDNA region, it is also worth noting that
P. muris had longer sequences (about 68bp at 5' end) than other species of the
Plagiorchis genus. The phylogenic tree of the family Plagiorchiidae is consistent with a previous molecular analysis of
Haematoloechus species and
Plagiorchis species using internally transcribed spacer 1 (ITS1), and with large subunit sequence data (
Snyder and Tkach, 2001). Tkach et al. (
2000a,
2000b,
2001) reported that the suborder Plagiorchiata is composed of two-supported clades, which can be considered superfamilies, namely, Plagiorchioidea including the Plagiorchiidae, and Microphalloidea based on partial lsr DNA sequences. In Plagiorchiidae, a close phylogenic relationship was observed between two
Plagiorchis species (
P. koreanus,
P. vespertilionis),
Lecithopyge rastellus, and
Haplometra cylindracea. Since there is a little data on mtCOI Plagiorchiidae worms, it is not possible to determine whether
P. muris is in the same clade as
Glypthelmins spp. However, we infer that
P. muris is probably in a separate clade (
Fig. 2). The present partial 28S rDNA sequence-base phylogenic analysis of the family Plagiorchiidae including
P. muris places the genera
Plagiorchis into a welldefined separate clade within the family Plagiorchiidae. The positions of the majority of the taxa of Plagiorchiidae are consistent with the traditional systematic views, the above molecular data also supports the traditional morphology-based conclusion that
P. muris belongs to
Plagiorchis spp.
Notes
-
Sequence data from this article have been deposited with the EMBL/GeneBank Data Libraries under Accession numbers 28S rDNA sequence (AF096222), mtCOI sequence (AF096236).
-
This study was supported by the research grant of Hallym University.
References
- 1. Chai JY, Lee SH. Intestinal trematodes infecting human in Korea. Southeast Asian J Trop Med Public Health 1991;22:163-170.
- 2. Garey JR, Wolstenholme DR. Platyhelminth mitochondrial DNA: evidence for early evolutionary origin of a tRNA ser AGN that contains a dihydrouridine arm replacement loop, and of serine-specifying AGA and AGG codons. J Mol Evol 1989;28:374-387.
- 3. Hills DM, Mortz C, Marble BK. Molecular Systematics. 1996, 2nd ed. Sunderland, USA. Sinauers Assoc. Inc.. pp 385-514.
- 4. Hong SJ, Woo HC, Lee SU, Huh S. Infection status of dragonflies with Plagiorchis muris metacercariae in Korea. Korean J Parasitol 1999;37:65-70.
- 5. Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980;16:111-120.
- 6. Littlewood DT, Bray RA. Interrelationships of platyhelminthes. 2001, London, U.K.. Tayler & Francis. pp 186-193.
- 7. Olson PD, Cribb TH, Tkach VV, Bray RA, Littlewood DT. Phylogeny and classification of the Digenea (Platyhelminthes: trematoda). Int J Parasitol 2003;33:733-755.
- 8. Qu LH, Nicoloso M, Bachellerie JP. Phylogenic calibration of the 5' terminal domain of large rRNA achieved by determining twenty eucaryotic sequences. J Mol Evol 1988;28:113-124.
- 9. Seo BS, Cho SY, Hong ST, Hong ST, Hong SJ, Lee SH. Studies on parasitic helminths of Korea. V. Survey on intestinal trematodes of house rats. Korean J Parasitol 1981;19:131-136.
- 10. Snyder SO, Tkach VV. Phylogenic and biogeographical relationships among some holarctic from lung flukes (Digenea: Haematoloechidae). J Parasitol 2001;87:1433-1440.
- 11. Swofford DL, Olsen GJ, Waddell PJ, Hillis DM. In Hillis DM, Moritz G, Mable BK eds, Phylogeny inference molecular systematics. 1996, 2nd Edition. Massachusetts. Sinauer Associates Sunderland. pp 407-514.
- 12. Tkach V, Grabda-Kazubska B, Swiderski Z. Systematic position and phylogenic relationships of the family Omphalometridae (Digenea, Plagiorchiida) inferred from partial 1srDNA sequences. Int J Parasitol 2001;31:81-85.
- 13. Tkach V, Pawlowski J, Mariaux J. Phylogenic analysis of the suborder Plagiorchiata (Platyhelminthes, Digenea) based on partial 1srDNA sequences. Int J Parasitol 2000a;30:83-93.
- 14. Tkach V, Pawlowski J, Sharpilo VP. Molecular and morphological differentiation between species of the Plagiorchis vespertilionis group (Digenea, Plagiorchiidae) occurring in European bats, with a re-description of P. vespertilionis (Muller, 1780). Syst Parasitol 2000b;47:9-22.
Fig. 1Phylogenic relationships between the 28S D1 rDNA gene of Plagiorchis muris and other Plagiorchiidae. This parsimonious tree was analyzed by neighbor-joining method using PHYLIP program. G, Glypthelmins; A, Astiotrema; H, Haplometra; L. rastellus, Lecithopyge rastellus; L. nigrovenosus, Leptophallus nigrovenosus; M. longicollis, Macrodera longicollis; N, Neoglyphe; P. cloacicola, Paralepoderma cloacicola; P, Plagiorchis; S, Skrjabinoeces; H. longiplexus, Haematoloechus longiplexus.

Fig. 2Phylogenic relationships of the mtCOI gene of Plagiorchis muris with other Plagiorchiidae. This parsimonious tree was analyzed by the neighbor-joining method using PHYLIP program. G, Glypthelmins; P. muris, Plagiorchis muris; P. macrorchis, Paragonimus macrorchis.
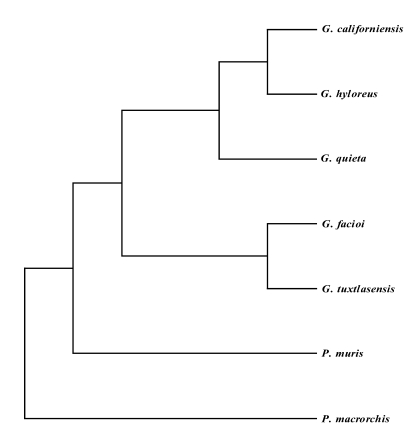
Table 1.Plagiorchiidae species used in this study, GenBank accession numbers for corresponding sequences, sequence lengths of the 28S D1 rDNA gene
Table 1.
|
Species |
GenBank No. |
Sequence length (bp) |
Aligned sequence length (bp)b)
|
Reference |
|
Astiotrema monticellii
|
AF184253 |
1,261 |
231 |
Littlewood and Bray, 2001 |
|
Glypthelmins californiensis
|
AY278052 |
1,275 |
222 |
|
|
G. facioi
|
AY278046 |
1,275 |
222 |
|
|
G. hyloreus
|
AY278050 |
1,274 |
222 |
|
|
G. pennsylvaniensis
|
AF433676 |
1,250 |
230 |
Tkach et al., 2001 |
|
G. quieta
|
AY222278 |
1,256 |
227 |
Olson et al., 2003 |
|
G. tuxtlasensis
|
AY278048 |
1,274 |
223 |
|
|
|
Haplometra cylindracea
|
AF151933 |
1,254 |
230 |
Tkach et al., 2000a, 2001 |
|
|
Lecithopyge rastellus
|
AF151932 |
1,254 |
230 |
Tkach et al., 2000a, 2001 |
|
Leptophallus nigrovenosus
|
AF151914 |
1,256 |
229 |
Tkach et al., 2000a |
|
Macrodera longicollis
|
AF151913 |
1,257 |
229 |
Tkach et al., 2000a |
|
Metaleptophallus gracillimus
|
AF151912 |
1,256 |
229 |
Tkach et al., 2000a |
|
Neoglyphe locellus
|
AF300330 |
1,255 |
230 |
|
|
N. sobolevi
|
AF300329 |
1,255 |
230 |
Tkach et al., 2001 |
|
Paralepoderma cloacicola
|
AF151910 |
1,255 |
229 |
Tkach et al., 2000a |
|
Plagiorchis eleganse
|
AF151911 |
1,263 |
230 |
Tkach et al., 2000b |
|
P. koreanus
|
AF151930 |
1,254 |
230 |
Tkach et al., 2000ab, 2001 |
|
P. muelleri
|
AF184250 |
1,254 |
230 |
Tkach et al., 2000b |
|
P. muris
|
AF096222a)
|
309 |
309 |
Present paper |
|
P. vespertilionis
|
AF151931 |
1,254 |
230 |
Tkach et al., 2000ab, 2001 |
|
Skrjabinoeces similis
|
AY222279 |
1,255 |
232 |
Olson et al., 2003 |
Table 2.Plagiorchiidae species used in this study, GenBank accession numbers for corresponding sequences, sequence lengths for the mtCOI gene
Table 2.
|
Species |
GenBank No. |
Sequence length (bp) |
Aligned sequence length (bp) |
Reference |
|
Glypthelmins californiensis
|
AY278058 |
381 |
381 |
– |
|
G. facioi
|
AY278053 |
383 |
383 |
– |
|
G. hyloreus
|
AY278059 |
381 |
381 |
– |
|
G. quieta
|
AY278056 |
381 |
381 |
– |
|
G. tuxtlasensis
|
AY278054 |
381 |
381 |
– |
|
Plagiorchis muris
|
AF096236a)
|
443 |
397 |
Present paper |
Citations
Citations to this article as recorded by

- Raccoons contraband – The metazoan parasite fauna of free-ranging raccoons in central Europe
Norbert Peter, Dorian D. Dörge, Sarah Cunze, Anna V. Schantz, Ajdin Skaljic, Sonja Rueckert, Sven Klimpel
International Journal for Parasitology: Parasites and Wildlife.2023; 20: 79. CrossRef - Trematode diversity in freshwater snails from a stopover point for migratory waterfowls in Hokkaido, Japan: An assessment by molecular phylogenetic and population genetic analyses
Minoru Nakao, Mizuki Sasaki
Parasitology International.2021; 83: 102329. CrossRef - Parasites Circulating in Wild Synanthropic Capybaras (Hydrochoerus hydrochaeris): A One Health Approach
Manuel Uribe, Carlos Hermosilla, Arlex Rodríguez-Durán, Juan Vélez, Sara López-Osorio, Jenny J. Chaparro-Gutiérrez, Jesús A. Cortés-Vecino
Pathogens.2021; 10(9): 1152. CrossRef - Plagiorchis sp. in small mammals of Senegal and the potential emergence of a zoonotic trematodiasis
Stefano Catalano, Steven A. Nadler, Cheikh B. Fall, Kirsty J. Marsh, Elsa Léger, Mariama Sène, Simon L. Priestnall, Chelsea L. Wood, Nicolas D. Diouf, Khalilou Bâ, Joanne P. Webster
International Journal for Parasitology: Parasites and Wildlife.2019; 8: 164. CrossRef - Characterization of the complete mitochondrial genome of Plagiorchis maculosus (Digenea, Plagiorchiidae), Representative of a taxonomically complex digenean family
Suleman, Jun Ma, Mian Sayed Khan, Vasyl V. Tkach, Nehaz Muhammad, Dong Zhang, Xing-Quan Zhu
Parasitology International.2019; 71: 99. CrossRef - Molecular analyses reveal high species diversity of trematodes in a sub-Arctic lake
Miroslava Soldánová, Simona Georgieva, Jana Roháčová, Rune Knudsen, Jesper A. Kuhn, Eirik H. Henriksen, Anna Siwertsson, Jenny C. Shaw, Armand M. Kuris, Per-Arne Amundsen, Tomáš Scholz, Kevin D. Lafferty, Aneta Kostadinova
International Journal for Parasitology.2017; 47(6): 327. CrossRef - Phylogenetic Relationships of 3 Korean Neodiplostomum Species (Digenea: Neodiplostomidae) Based on Partial CO1 Gene
Kyoung-Ho Pyo, Jo Woon Yi Lee, Jin-Ju Lee, Yun-Kyu Park, Eun-Hee Shin, Jong-Yil Chai
The Korean Journal of Parasitology.2014; 52(3): 325. CrossRef - A molecular and ecological analysis of the trematodePlagiorchis elegansin the wood mouseApodemus sylvaticusfrom a periaquatic ecosystem in the UK
K. Boyce, G. Hide, P.S. Craig, C. Reynolds, M. Hussain, A.J. Bodell, H. Bradshaw, A. Pickles, M.T. Rogan
Journal of Helminthology.2014; 88(3): 310. CrossRef - The life cycle of Asymphylodora perccotti sp. n. (Trematoda: Lissorchiidae) in the Russian Southern Far East
Vladimir V. Besprozvannykh, Alexey V. Ermolenko, Dmitry M. Atopkin
Parasitology International.2012; 61(2): 235. CrossRef - Life cycle of Renylaima capensis, a brachylaimid trematode of shrews and slugs in South Africa: two-host and three-host transmission modalities suggested by epizootiology and DNA sequencing
Wilhelm F Sirgel, Patricio Artigas, M Dolores Bargues, Santiago Mas-Coma
Parasites & Vectors.2012;[Epub] CrossRef - The occurrence of the trematodePlagiorchis murisin the wood mouseApodemus sylvaticusin North Yorkshire, UK
M.T. Rogan, P.S. Craig, G. Hide, S. Heath, A. Pickles, D.M. Storey
Journal of Helminthology.2007; 81(1): 57. CrossRef - Molecular phylogeny of parasitic Platyhelminthes based on sequences of partial 28S rDNA D1 and mitochondrial cytochrome c oxidase subunit I
Soo-Ung Lee, Ha-Chung Chun, Sun Huh
The Korean Journal of Parasitology.2007; 45(3): 181. CrossRef