Molecular Phylogeny of Acanthamoeba
Article information
Abstract
After morphological grouping of Acanthamoeba by Pussard and Pons, phylogeny of the genus has been always a big topic to the researchers. Because of the variability of morphological characteristics, unchangeable and stable characters have been investigated for phylogenic criteria. Isoenzyme and mitochondrial DNA RFLP (Mt DNA RFLP) analyses revealed different patterns among strains assigned to a same species. Therefore, these characteristics would be considered as tools for strain discrimination than species identification. The most recently developed and the most promising method is the sequence analysis of 18s ribosomal RNA coding DNA (18s rDNA). The phylogenic tree based on comparison of 18s rDNA sequences distinguished the 3 morphological groups of Acanthamoeba and divided them into 12 unique sequence types (T1-T12 genotypes). Most clinical and environmental isolates belonged to the morphological group II and the genotype T4. In the Republic of Korea, 2 strains of Acanthamoeba, YM-2 and YM-3, were first isolated from the environment in 1974. However, phylogenic identification of Korean Acanthamoeba isolates from human infections or the environment were tried from the late 1990s. By RFLP analysis or total sequence analysis of 18s rDNA revealed that almost all clinical isolates including the one from a suspicious granulomatous amebic encephalitis patient belonged to the genotype T4. A large number of environmental isolates from contact lens storage cases, tapped water, and ocean sediments also belonged to the genotype T4. Almost identical strain characteristics, such as Mt DNA RFLP pattern of environmental isolates, with the clinical isolates could make a simple conclusion that most environmental isolates might be a potential keratopathogen.
INTRODUCTION
The genus Acanthamoeba Volkonsky, 1931 has adapted to live in a variety of human environments, such as soil, air, freshwater, swimming pools, tap water, and ocean sediments. Recently, medical protozoologists have paid attention to Acanthamoeba since several species of the genus have been known to cause life-threatening granulomatous amebic encephalitis (GAE) in immunocompromised individuals and sight-threatening amebic keratitis in contact lens wearers. The incidence of Acanthamoeba keratitis infection has increased exponentially during the last 30 years [1,2]. Up to the year 2009, 2 suspicious GAE cases were found to be infected with Acanthamoeba spp. in the Republic of Korea [3,4]. These patients died of meningoencephalitis. In addition, 1 case of Acanthamoeba pneumonia with an immunodeficiency status was reported [4]. In the case of Acanthamoeba keratitis, the incidence is increasing from the early 1990s and more than 30 cases were reported already.
Despite the medical importance of the genus, identification and classification of acanthamoebae at the subgenus level has been problematic. The identification of Acanthamoeba at the generic level can be easily accomplished by the morphological characteristics. Pussard and Pons [5] divided the genus into 3 morphological groups according to the cyst size and other morphological features. Group 1 consists of Acanthamoeba spp. with relatively large cysts, distinctly stellate endocysts, and smooth spherical ectocysts. Group 2 and group 3 Acanthamoeba spp. have smaller cysts (less than 18 µm in diameter). Species in Group 2 have polygonal to stellate endocysts with irregular or wrinkled ectocysts, while the cysts of group 3 species have rounded or slightly angular endocysts with tinner and smooth or slightly wrinkled ectocysts. The grouping has been widely used before species identification of the amoeba. However, the morphologic characteristics of the cyst can change with the culture conditions [6,7] and highly variable within the same strain (Fig. 1). Therefore, species identification by morphology alone can hardly be possible [8].
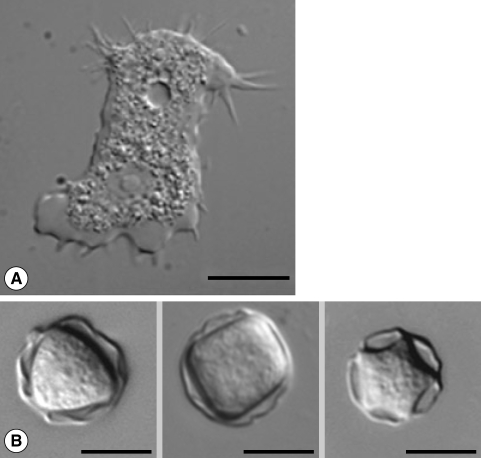
Microphotographs of Acanthamoeba. (A) Trophozoite of A. castellanii Castellani with many characteristic pin-like acanthopodia and numerous vacuoles. Nucleus and a contractile vacuole are prominent. (B) Various shaped cysts of Acanthamoeba with different arm numbers.
Investigators have used several kinds of non-morphological methods for the taxonomy of Acanthamoeba spp. Among the methods, isoenzyme analysis, RFLP of mitochondrial DNA (Mt DNA), RFLP of small subunit ribosomal DNA (ssu rDNA), and the sequence analysis of ssu rDNA have been recently applied and regarded promising. In this paper, the potential of these molecular methods for the phylogeny of Acanthamoeba, and the molecular phylogeny of Acanthamoeba korean isolates, were briefly reviewed.
ISOENZYME ANALYSIS AND MITOCHONDRIAL DNA RFLP
Isoelectric focusing of isoenzymes had been applied for identification of Acanthamoeba spp. Studies of isoenzyme patterns [9-12] suggested the other groupings of Acanthamoeba strains that are not consistent with previous species assignment based on the morphological criteria of Pussard and Pons [5]. De Jonckheerez [13] examined 30 strains of 15 Acanthamoeba species for the isoenzyme patterns by agarose isoelectric focusing (IEF) in the pH range 3-10. Comparing zymograms of 5 enzymes, he reassigned the strains of amoebae to each species. Kong et al. [14] observed the interstrain polymorphism of the isoenzyme profiles among 7 strains of Acanthamoeba morphologically assigned to Aanthamoeba polyphaga. Since considerable variation of zymograms was noted within a species of Acanthamoeba, isoenzyme study alone is not regarded fully sufficient for species identification.
Mt DNA RFLP analysis has also been applied for taxonomy of Acanthamoeba. Bogler et al. [15] prospected the availability of Mt DNA RFLP for the identification of Acanthamoeba after analysis of 13 strains. They observed that most of the Acanthamoeba strains had distinct fragment patterns of Mt DNA. Yagita and Endo [16] also analyzed 8 isolates which had been identified as Acanthamoeba castellaniii or A. polyphaga, and reported variability of Mt DNA digestion phenotypes among different strains of A. castellanii. Kong et al. [14] confirmed the variability in digestion phenotype of A. polyphaga as well as that of A. castellanii. As for the interstrain sequence diversity of Mt DNA of Acanthamoeba, other investigators reported similar results [15,17-19].
Considering profound interstrain diversity in a species of the Mt DNA RFLP and alloenzyme profiles, Kong et al. [14] and Chung et al. [20] suggested that both analyses should be used for strain identification, differentiation, and characterization rather than species identification.
SMALL SUBUNIT RIBOSOMAL RNA CODING DNA RFLP (RIBOPRINTING)
PCR-based DNA restriction analyses of 18s rDNA (riboprinting) of Acanthamoeba showed promising results for species differentiation of Acanthamoeba [21]. The sequence of primers for PCR was designed to hybridize at the highly conserved sequences at the 5'and 3'termini of eukaryotic ssu rDNA [22]. They analyzed 23 reference strains of Acanthamoeba, including 18 (neo) type strains for classification at the subgenus level by riboprinting. The dendrogram based on the riboprinting coincided well with the grouping of Pussard and Pons [5] based on the morphological features of the cysts. Khan and Paget [23] applied similar methods to identify clinical and environmental isolates from UK. They amplified a part of 18S rDNA with genus-specific primers [23] and digested the products with restriction enzymes. The riboprinting can be applied for rapid identification of Acanthamoeba isolated from the clinical and environmental specimens under limited condition to obtain the DNA sequence of the samples.
18S rDNA SEQUENCE ANALYSIS
The most recently proposed method for molecular taxonomy of Acanthamoeba is 18s rDNA sequence analysis. Byers group investigated the 18s rRNA gene phylogeny [24,25]. After the examination of the 18s rDNA sequence variation in a group of 53 strains that includes the morphologically assigned 18 isolates and 35 new isolates, they suggested 12 sequence types (T1-T12). The sequence types were defined as the sequences or groups of sequences that differ from all other sequences by at least 6%. In the phylogeny by 18s rDNA sequence analysis, 3 morphological groups were clearly distinguished. Each of the morphological groups consisted of several different lineages of sequence types. Three species in the morphological group I were identified as T7 for Acanthamoeba astronyxis, T8 for Acanthamoeba tubiashi, and T9 for Acanthamoeba comandoni. In group 3, Acanthamoeba culbertsoni was identified as T10 and A. healyi as T12, and Acanthamoeba palestinensis, Acanthamoeba pustulosa, and Acanthamoeba lenticulata represented T1, T2, and T5, respectively. The other 3 types, T3, T4, and T11 are included in group 2; T3 for Acanthamoeba griffinii and T11 for Acanthamoeba hatchetti and Acanthamoeba stevensoni. The biggest lineage T4 contains all the remaining group 2 isolates. However, by this system, the relationship among all taxa within T4 was not clearly defined. Strains in T4 are too closely related to be resolved fully by use of 18s rDNA sequences. They proposed that each sequence type should be equated with a single species. The various species in T4 might be all reclassified as A. castellanii, because the sequence type includes the type strain for that species.
Although phylogeny by 18s rDNA sequence analysis has a limitation to distinguish species in sequence type T4, it is practical to classify clinical or environmental samples for species identification. Now the identification of a new isolate of Acanthamoeba from clinical sample or environment is performed mainly by 18s rDNA sequence analysis [26-29].
With the 18s rDNA sequence analysis, reassignment of several Acanthamoeba spp. originally assigned by morphological criteria have been suggested. Chung's group evaluated the taxonomic validity of 4 species of Acanthamoeba; A. divionensis, A. paradivionensis, A. mauritaniensis, and A. rhysodes and suggested that 3 species should be regarded as synonyms of A. rhysodes. They exhibited 18s rDNA sequence differences of 0.2-1.1 with each other.
Since most clinical or environmental isolates belong to 18s rDNA sequence type T4, many investigators prefer to classify an isolate as their genotypes than scientific names even in the case of already named species, such as A. castellanii Castellani. It is necessary and urgent to define the strains in the genotype T4 as a species complex or as several species based on other molecular characteristics.
MITOCHONDRIAL 16S rDNA SEQUENCE ANALYSIS
The value of mitochondrial small subunit rRNA gene (mt ssu rDNA) analysis as a taxonomic tool for Acanthamoeba isolates with close interrelationship was also evaluated [30,31]. Mt DNA is a molecular clone within mitochondriate eukaryotic cells like Acanthamoeba. Merit of Mt ssu rDNA is lack of intron, and smaller and more consistent in size than nuclear ssu rDNA. Yu et al. [30] compared the phylogeny based on the PCR-RFLP pattern of mt ssu rDNA with that of genomic ssu rDNA PCR-RFLP. Ledee et al. [31] analyzed Mt ssu rDNA sequences of 68 isolates and constructed a phylogenetic tree. The similar genotype clustering by Mt ssu rDNA analysis with that by nuclear ssu rDNA genes indicated that those clustering represent the main branches in the phylogeny of Acanthamoeba. Moreover, based on the subgrouping of T4 genotypes into 8 clades, they suggested that Mt ssu rDNA sequence may serve as the best available molecular basis for differentiating the species with this genotype.
Acanthamoeba PHYLOGENY IN KOREA
It was middle of the 1970s that the first suspicious Acanthamoeba meningoencephalitis case was reported in the Republic of Korea [3]. The patient was a 5-year-old boy who developed multiple subcutaneous, non-tuberculous granulomas in many organs, and died of meningoencephalitis. However, unfortunately the organism identification was not performed. It was also in the middle of the 1970s that the first strain of Acanthamoeba was isolated in Korea [32]. Acanthamoeba spp., YM-2, YM-3, and YM-4, were isolated from water puddles, a storing reservoir, and the gill of fresh water fish. However, the phylogenic positioning of those isolates was tried quite long time later because of lack of standardized species identification system.
Molecular phylogeny of clinical isolates
The second case of Acanthamoeba meningoencephalitis in Korea was a 7-year-old boy with facial laceration near eyebrow by accident. Acanthamoeba trophozoites were observed in his skin biopsy after his death [4]. However, species identification of the causative Acanthamoeba was tried after 13 years later based on 18s rDNA partial sequence analysis. For partial amplification, 2 sets of genus-specific primers were designed to hybridize to the specific regions of 18S rDNA of the genus Acanthamoeba (Fig. 2). The primer sets, VR1 and VR2, encompassing the variable regions of A. castellanii Castellani 18S rDNA were designed to hybridize from 111-326 and 822-1,090 nucleotide numbers, respectively. The sequences of these primers were as follows: VR1F (TTATAGTTTATTTGATGGTC), VR1R (TGCGATCCGAAAGA-GTTACT), VR2F (CCGTCCCCTCCTTCTGGATT) and VR2R (CTATCCCTATT-AATCATTAC). Two fragments, sized 200 and 270 bp, of the variable region of 18s rDNA was amplified by PCR from the paraffin block of the skin biopsy. Combined sequences of 2 variable regions were subjected to construct phylogenetic tree by comparison with that of reference strains (Fig. 3). The causative organism of the case was identified as A. castellanii complex (T4 genotype) and was genetically closely related to 2 clinical isolates. AY026243 is A. polyphaga Jones strain, the first Acanthamoeba keratitis isolate [33], and the other strain U07410 is also isolated from a keratitis patient in the USA.
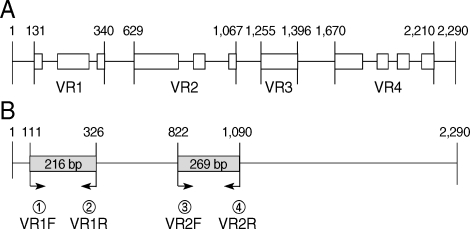
Schematic diagram of 18s rDNA sequence of A. castellanii Casteallani. (A) 4 variable regions of 18s rDNA are demonstrated with white boxes and named as VR1, VR2, VR3, and VR4. (B) Primer sets for PCR of 2 parts of variable regions are presented.
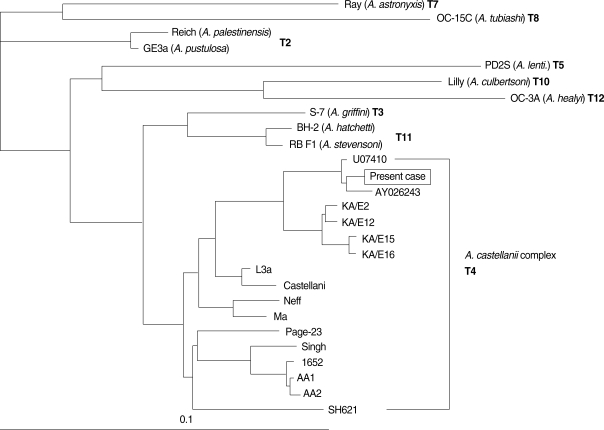
Phylogenetic dendrogram derived by aligning the partial 18s rDNA from the present case and various strains of Acanthamoeba in the database. U07410 and AY026243 are Genbank accession numbers of 18s rDNA sequences of the 2 Acanthamoeba isolates.
Four Acanthamoeba isolates (KA/E2, KA/E12, KA/E15 and KA/E16) from infected cornea of Korean patients were reported as Acanthamoeba lugdunensis based on the Mt DNA RFLP and riboprinting of 18s rDNA [34]. Three isolates, with the exception of KA/E15, revealed the identical Mt DNA RFLPs and riboprint patterns with Acanthamoeba L3a, the type strain of A. lugdunensis. Identification of the isolates as A. lugdunensis was confirmed by 18s rDNA sequence analysis. The sequence difference of the 4 isolates from L3a strain was 0.1% to 0.4%, and that from A. castellanii Castellani was 1.2% to 1.5%.
Amebic keratitis related to orthokeratology lens overnight wear was reported in a 15-year-old girl who wore the lens for 10 months [29]. Based on the 18s rDNA sequence analysis, the infected Acanthamoeba was identified as the genotype T4 and very closely related to A. castellanii Ma strain which had been isolated also from amoebic keratitis.
Xuan et al. [35] characterized 3 Acanthamoeba isolates (KA/E9, KA/E17, and KA/E23) from Korean keratitis patients and identified as Acanthamoeba triangularis by the 18s rDNA sequence homology. The 3 isolates showed 1.3-1.6% of 18s rDNA sequence differences from A. triangularis. Although the cyst morphology of the 3 isolates was not identical with A. triangularis, their mitochondrial DNA RFLP was very similar with that of A. triangularis.
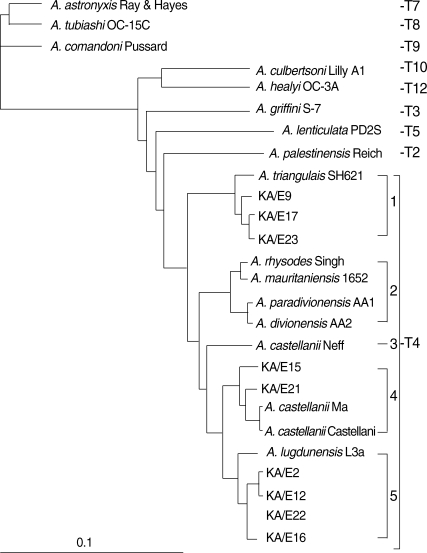
Here we present the phylogenetic tree of Acanthamoeba by 18s rDNA sequence analysis of 22 known strains as reference strains and 10 clinical isolates (KA/E series) from Korean keratitis patients (Table 1 & Fig. 4). All clinical isolates from Korean patients belonged to the morphological group II and identified as the sequence type T4. They scattered into 4 subclades within T4. It is not certain to assign them as separate species or A. castellanii species complex. By Mt 16S rDNA sequence analysis of Korean keratitis isolates, the strains in the T4 genotype were subgrouped into 5 clades (Fig. 5). A. castellanii Castellani and A. lugdunensis L3a belonged to different clades. With these results, we confirmed the potential of Mt 16s rDNA to identify species in genotype T4 as suggested by Ledee et al. [31].
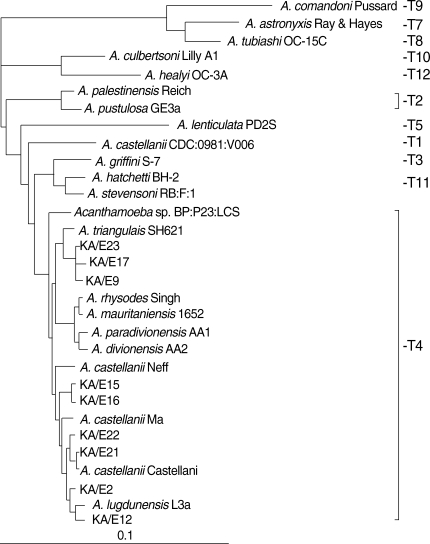
Neighbour-joining dendrogram based on comparative 18s rDNA sequence analysis showing relationships of Acanthamoeba isolates from Korean keratitis patients to representative members of the genus Acanthamoeba.
Molecular phylogeny of environmental isolates
Acanthamoeba isolate from Korean soil with strain name KA/S2 was characterized by Chung et al. [20]. Mt DNA RFLP patterns of KA/S2 were identical with those of the Neff strain originally isolated from the soil of the USA.
By a nationwide survey on the contamination rate of Acanthamoeba in contact lens storage cases, 110 Acanthamoeba isolates were identified and characterized by comparison of riboprinting and Mt DNA RFLP with those of reference strains obtained from American Type Culture Collections (ATCC) [36-38]. All isolates from the lens storage cases belonged to the morphological group II and were identified as the genotype T4. Their individual genetic characteristics by Mt DNA RFLP showed that the most common pattern was that of A. lugdunensis L3a and KA/E2, the type stain and a corneal isolate from a Korean keratitis patient, respectively. Another common pattern was that of A. castellanii Castellani and KA/E3, the type strain and another corneal isolate found in Korea. The pattern of A. castellanii Ma strain, a corneal isolate from the USA was also quite common in some part of Korea. The prevalent pattern for each type of Acanthamoeba in southwestern area was very different from that in southeastern areas and Seoul, Korea [38]. Most Acanthamoeba isolates from domestic tap water were identified as potential keratopathogens based on the molecular characteristics of Mt DNA RFLP and 18s rDNA sequence analysis [39]. Interestingly, the Mt DNA RFLP patterns of 5 isolates from contact lens storage cases were found to be the same as those of isolates from the lens owner's domestic tap water.
Acanthamoeba isolates from ocean sediments of Korea showed big diversity in genetic characteristics compared to the isolates from contact storage cases [40]. All of 18 isolates belonged to the morphologic group II. Fifteen isolates showed close relatedness with clinical isolates and A. castellanii Castellani, and they formed a lineage equivalent to the T4 genotype. Two reference strains from the ocean sediment, A. hatchetti BH-2 and A. griffini S-7, clustered unequivocally with these 15 isolates.
Im and Shin reported [41] an isolate from a freshwater fish in Korea as a new species of Acanthamoeba and assigned as Acanthamoeba sohi, based on random amplified polymorphic DNA marker analysis, Mt DNA RFLP, and 18s rDNA sequence analysis [41]. The isolate belonged to the morphologic group III and formed a different clade from 3 reference strains of group 3, A. culbertsoni, A. healyi, and Acanthamoeba royreba in the phylogenic tree.
CONCLUSION
Although various molecular characteristics have been applied for the taxonomy of Acanthamoeba up to date, hitherto species identification system needs to be improved. Sequence data of 18s rDNA may provide information for genotyping or grouping to the most closely related known species. Mt 16s rDNA sequence analysis could be an additive data to subgroup the strains in the genotype T4, to which most clinical and environmental isolates belong. More detailed molecular characteristics could be obtained by isoenzyme analysis and Mt DNA RFLP. Comparison of molecular characteristics of clinical or environmental isolates with already identified strains is also a short way to identify a new isolate.
ACKNOWLEDGEMENTS
The author thanks Prof. Dong-Il Chung, Kyungpook National University School of Medicine, for careful reading of this manuscript and helpful discussions.