Giardia intraflagellar transport protein 88 is involved in flagella formation
Article information
Abstract
Intraflagellar transport (IFT) particles, a multi-protein apparatus composed of complex A and B, are known to be involved in homeostasis of flagella formation. IFT particles have recently become an interesting topic in Giardia lamblia, which has 4 pairs of flagella. In this experiment, we examined the function of giardial IFT components. When 7 components (IFT121, 140, 20, 46, 52, 81, and 88) of IFT were expressed in Giardia trophozoites as a tagged form with mNeonGreen, all of them were found in both flagella pores and cytoplasmic axonemes. In addition, motor proteins for IFT particles (kinesin-13 and kinesin-2b), were localized to a median body and cytoplasmic flagella, respectively. The CRISPRi-mediated knockdown of IFT88 significantly affected the lengths of all 4 flagella compared to the control cells, Giardia expressing dead Cas9 using control guide RNA. Decreased expression of kinesin-2b also resulted in shortening of flagella, excluding the ventral flagella. Live Giardia cells expressing IFT88-mNeonGreen clearly demonstrated fluorescence in flagella pores and cytoplasmic axonemes. These results on IFT88 and kinesin-2b indicate that IFT complex plays a role in maintenance of G. lamblia flagella.
Introduction
Flagella and cilia are microtubule (MT)-based organelles involved in diverse functions of eukaryotic cells such as motility, attachment, signaling responses to chemicals, cell division, and development [1]. The basic structure for these appendages is highly conserved axoneme which is comprised of 9 MT doublets with a central MT pair [2]. The dynamics and maintenance of these structures are dependent upon bidirectional intraflagellar transport (IFT) of MT building blocks to the growing ends of flagella and cilia [3]. IFT particles, the main components of IFT train, consist of 2 complexes, complex A and complex B, which have differential roles for retrograde and anterograde transports of flagellar biogenesis [4]. Specific motor proteins are known for bidirectional IFTs, i.e., dynein for retrograde transport and heterotrimeric kinesin-2 (kinesin-2a, kinesin-2b, and kinesin associated protein) for anterograde transport [4]. Kinesin-13 was discovered as a depolymerizer of MTs, contributing to the equilibrium of ciliary length in Xenopus [5]. In addition, a heterooctameric protein responsible for Bardet-Biedl syndrome, BBSome, was found as a cargo adaptor involved in homeostasis of flagella and cilia [6].
Giardia lamblia is an interesting protozoan with 4 bilaterally symmetric pairs of flagella that differ in their intracellular locations and lengths. Unlike many eukaryotic organism, Giardia lacks a transition zone (TZ) at the flagella and does not have TZ proteins [7]. However, this organism has IFT homologs, motor proteins, and some BBSome components in its genome [8]. A study using live-cell quantitative imaging and mathematical modeling of IFT components and kinesin-13 indicated a model in which length of each pair of flagella is coordinated by length-independent IFT-mediated assembly and length-dependent kinesin-13-mediated disassembly of flagella [9]. Role of kinesin-13 in this control was proven by knockdown (KD) of kinesin-13 expression using dead Cas9 (dCas9), resulting in increased flagella length [9,10]. Decreased expression of kinesin-2a via CRISPRi-mediated KD produced Giardia cells with shortened flagella, demonstrating that the kinesin-2a function as a motor protein for the anterograde IFT complex B [10]. Interestingly, biogenesis of 4 pairs of Giardia flagella was differentially related to production of other organelles [11]. Formation of marginal plate occurs in relation with the anterior axonemes whereas a structure called funis is connected with membranous caudal flagella [12,13]. In addition, multiplication and repositioning of flagellar basal bodies are pre-requisite for cell division [14] and flagella-mediated motility is required for cytokinesis instead of actin-myosin function [15].
To establish a system for observing flagellar formation in G. lamblia, we generated several cells expressing mNeonGreen (mNG)-tagged IFTs and examined for their localization. The roles of IFT88, a putative component of IFT complex B, and its cognate motor protein, kinesin-2b, were examined using CRISPRi-mediated KD experiments.
Materials and Methods
Ethics statement
Not applicable.
Giardia lamblia culture conditions
G. lamblia trophozoites (Strain WB, ATCC30957) (American Type Culture Collection, Manassas, VA, USA) were cultured in modified TYI-S-33 medium (2% casein digest, 1% yeast extract, 1% glucose, 0.2% NaCl, 0.2% cysteine, 0.02% ascorbic acid, 0.2% K2HPO4, 0.06% KH2PO4, pH 7.1) supplemented with 0.75 mg/ml bovine bile and 10% heat-inactivated calf serum (Gibco, Rockville, MD, USA) at 37°C [16]. Transfected strains were maintained in TYI-S-33 medium with antibiotics at the following concentrations (50 μg/ml puromycin and 600 μg/ml G418).
Construction of Giardia strain expressing C-terminal mNG tagged proteins
Several IFTs (IFT20, 46, 52, 81, 88, 121, and 140), kinesin-2b, and kinesin-13 homologs were identified by homology searches in the Giardia genome using GiardiaDB (https://giardiadb.org/giardiadb/app). Gene IDs of all isolated IFT components and motor proteins from GiardiaDB are described in Supplementary Table S1. To make C-terminal mNG tagged construct, mNG gene was amplified from mNG mRuby2-FRET-10 plasmid (Addgene, Watertown, MA, USA, Cat. No. 58179) by PCR using 2 primers, N11-linker-F-BamH1 and mNG-R-ER1 (Supplementary Table S2). The resulting fragment was cloned into the BamHI and EcoRI sites of pKS-3HA.NEO [17] to obtain pKS-N11-mNG.NEO (Supplementary Table S1). All IFT, kinesin-2b, and kinesin-13 genes were amplified from Giardia genomic DNA (gDNA) by PCR using appropriate primers (Supplementary Table S2), and the DNA fragments were then cloned into pKS-N11-mNG.NEO vector to generate constructs encoding the C-terminus with mNG tag. The list of plasmid in this study is described in Supplementary Table S1. All constructs were verified by DNA sequencing (Macrogen, Seoul, Korea). The established plasmids were linearized for integration into the native locus of the chromosome using its unique restriction site (the enzyme sites for each gene used were as follows: IFT20, HindIII; IFT46, NheI; IFT52, MluI; IFT81, AflII; IFT88, BsiWI; IFT121, XbaI; IFT140, BglII; kinesin-2b, BamHI; kinesin-13, NdeI) as described [18]. Briefly, the resulting fragments were gel purified using a PCR/gel combo kit (NucleoGen, Seoul, Korea) and 10 μg of the linearized DNA was used for transfection. Introduction of the DNA into Giardia trophozoites was performed by electroporation under the following conditions: 350 volts, 1,000 μF, and 700 Ω (BioRad, Hercules, CA, USA). The gDNA was then extracted from the resutlig transfectants using GeneAll GenEx kit (GeneAll Biotechnology, Seoul, Korea), and the extracted gDNA was used as templates to confirm whether the C-terminal mNG tag gene of IFT was integrated into the genome by PCR amplification using appropriate primer sets (Supplementary Table S2).
Fixed cell preparation
Giardia trophozoites were grown to 90% confluency. To harvest cells, culture tubes were incubated on ice for 15 min, and centrifuged at 1,900 g at 4°C for 15 min. The cell pellets were washed three times with ice cold 1× phosphate buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.4). Cells were fixed with PBS containing 2% paraformaldehyde, 100 mM 3-maleimidobenzoic acid N-hydroxysuccinimide ester (Sigma-Aldrich, St. Louis, MO, USA), and 100 mM ethylene glycol-bis(succinimidyl succinate) (ThermoFisher Scientific, Waltham, MA, USA) for 30 min at room temperature (RT). The cells were attached on the poly-L-lysine coated glass slides for 10 min, and then permeabilized with 0.5% Triton X-100 in PBS for 10 min at RT. After three washes with PBS, the slides were mounted with ProLong antifade with 4,6-diamino-2-phenylindole (Invitrogen, Carlsbad, CA, USA) and observed under an Axiovert 200 microscope (Carl Zeiss, Oberkochen, Germany).
dCas9-mediated KD
A plasmid containing dCas9, an endonuclease deficient Cas9, and guide RNA (gRNA)-expression cassette was synthesized based on a previous publication [10] (Macrogen, Seoul, Korea), and the resulting plasmid was named pdCas9.PAC (Supplementary Table S1). Specific gRNAs for each gene were designed by CRISPR RGEN Tools (http://www.rgenome.net/cas-designer/) using a NGG PAM sequence based on the G. lamblia ATCC 50803 genome (GenBank Assembly GCA_000002435.1). The list of oligonucleotides encoding ift88, kinesin-2b, and a control gRNA were described in Supplementary Table S2. Using these primers, double stranded gRNAs were synthesized and cloned into the BbsI site of pdCas9.PAC. IFT88 gRNAs targeted the nucleotides corresponding to +33, +258, and +455 of the IFT88 coding region, and kinesin-2b gRNAs aimed to the bases corresponding to +21, +141, and +271 of the kinesin-2b open reading frame. The resulting gRNA expressing plasmids were pdCas9-IFT88-g33, pdCas9-IFT88-g258, pdCas9-IFT88-g455, pdCas9-Kin2b-g21, pdCas9-Kin2b-g141, and pdCas9-Kin2b-g271. A control gRNA expression plasmid, pdCas9-gCont, was used to compare the effect of IFT88 and kinesin-2b KD cells (Supplementary Table S1).
Quantitation of transcriptional KD using quantitative PCR
Total RNA was prepared from Giardia carrying dCas9 and control gRNA, IFT88 gRNAs, or kinesin-2b gRNAs using TRIzol (ThermoFisher Scientific) according to the manufacturer’s instructions. Five micrograms of RNA were converted into complementary DNA using the Improm-II Reverse Transcription System (Promega, Madison, WI, USA). The quantitative PCR was performed using the LightCycler 480 SYBR Green I Master Kit (Roche Applied Science, Basel, Switzerland). The conditions for real-time PCR were as follows: pre-incubation for 5 min at 95°C and 45 cycles of 30 sec at 94°C, 30 sec at 56°C, and 30 sec at 72°C. The nucleotide sequences of the forward and reverse primers used for real-time quantitative PCR are listed in Supplementary Table S2. Transcript of G. lamblia actin-related gene (GiardiaDB; GL50803_15113) was used to normalize the amount of mRNA in the samples [19]. Relative quantifications of these data were determined by measuring the crossing-point value using the Light Cycler 480 II real-time PCR system software program (version LSC480 1.5.0.39, Roche Applied Science). Data were expressed as means± standard deviations of three independent experiments. All experiments were performed with three separate cultures.
Flagellar length measurements
Giardia cells were attached to slides, air-dried, and fixed with 100% methanol for 10 min. Then they were stained with 10% Giemsa solution (Sigma-Aldrich) for 10 min and washed with distilled water. After mounting with dibutyl phthalate xylene (Sigma-Aldrich), the cells were observed with an Axiovert 200 microscope (Carl Zeiss), and their differential interference contrast images were used to analyze the flagellar length. Each of 4 types of membrane-bound flagellar length were measured using the line freehand tracing mode in ImageJ software (http://imagej.nih.gov/ij/). Data on flagellar length were obtained from 35 cells per condition. Data are presented as the mean±standard deviation of three independent experiments.
Live-cell imaging of IFT88 in Giardia
After Giardia cells were incubated on ice for 15 min, they were harvested by centrifugation at 1,900 g for 15 min at 4°C. Cell pellets were washed 3 times in 5 ml of ice-cold 1× HEPES buffered saline (HBS; 137 mM NaCl, 5 mM KCl, 0.91 mM Na2HPO4, 5.55 mM glucose, and 20 mM HEPES, pH 7.0). The cells were then resuspended in 1 ml of 1× HBS. For live imaging, 500 μl of washed trophozoites were added to the center of a 27 mm glass base dish (Cat No. 150682, ThermoFisher Scientific) and incubated in a GasPak EZ anaerobic pouch (BD Biosciences, San Jose, CA, USA) for 1 h at 37°C. The dish was washed twice with warmed 1× HBS to remove unattached cells. Attached cells were overlaid with 2 ml of 2% ultra-low gelling agarose (A2576, Sigma-Aldrich) melted in 1× HBS containing a final concentration of 5 μM DRAQ5 (ThermoFisher Scientific) to stain the nuclei, and then left at RT for 30 min to solidify the agarose. Live imaging acquisition was performed on Elyra7 with Lattice SIM2 using a 63×1.4 NA under 5% CO2 at 37°C (Carl Zeiss). Data were processed using ZEN software (Blue Edition 3.0, Carl Zeiss).
Results
Localization of IFT and motor proteins in Giardia
In order to investigate the intracellular positions of the putative IFT components in Giardia trophozoites, 2 proteins (IFT121 and IFT140) and 5 proteins (IFT20, IFT46, IFT52, IFT81, and IFT88) were chosen as representatives for IFT complex A and B, respectively. In addition, 2 motor proteins, kinesin-13 and kinesin-2b were included in this study. All of these IFT genes were cloned into a plasmid in which they were expressed with a fluorescence tag, mNG from their own promoter (Fig. 1A). Upon transfection of these linearized plasmids into Giardia trophozoites, these constructs were integrated into Giardia chromosome (Fig. 1B). Integration of mNG-tagged IFT gene into the chromosome were confirmed by PCR using a specific primer for the target IFT and a reverse primer annealed to the neomycin gene. As shown in Fig. 1C, integration of IFT121- and IFT140-mNG resulted in PCR products of the expected sizes. On the other hand, PCR on Giardia control strain, WB with the identical primers did not make any DNA product. A nonspecific DNA fragment was generated in the PCR with Giardia cells with IFT140-mNG as indicated with an asterisk.
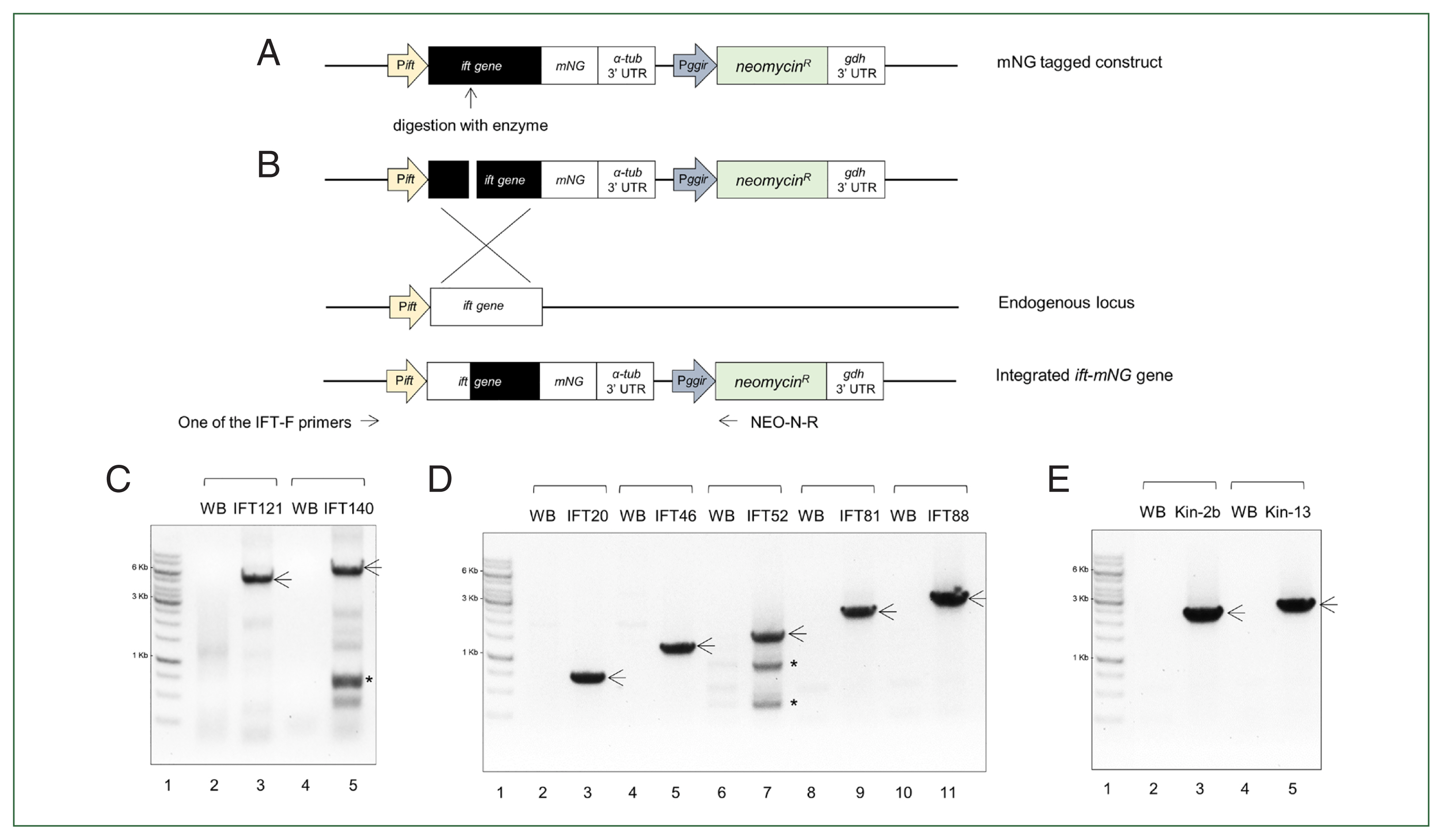
A schematic strategy and confirmation of the integration of intraflagellar transport (ift)-mNeonGreen (mNG) genes into Giardia genomic DNA (gDNA). (A) A schematic diagram of the construct of a full-length ift gene fused with an mNG tag based on a neomycin resistance cassette-containing plasmid. Pift, the promoter of each ift; α-tub 3′ UTR, 3′ untranslated region of Giardia α-tubulin (GL508003_103676); Pggir, the promoter region of Giardia giardin gene; gdh 3′ UTR, 3′ untranslated region of Giardia glutamate dehydrogenase (GL50803_21942); R, resistance. The open reading frame number of the IFTs for construct is as follows: 2 IFT-A components, IFT121 (GL508003_87817) and IFT140 (GL50803_17251), 5 IFT-B components, IFT20 (GL50803_3581), IFT46 (GL50803_7664), IFT52 (GL50803_0040995), IFT81 (GL50803_15428), and IFT88 (GL50803_0016660), 2 motor-related proteins, kinesin-2b (Kin-2b, GL50803_16456), kinesin-13 (Kin-13, GL50803_16945). The arrow indicates the enzyme site in each mNG-tagged construct to make a linearized form. (B) As schematic diagram of the incorporation of the ift-mNG gene into endogenous gDNA via homologues recombination. Arrows indicate the positions of primers used to confirm the integration of the constructs. The primer specific to each ift gene was designed as a forward primer, and then used along a reverse primer annealing to the neomycin resistance gene. (C) PCR analysis of IFT complex A-related genes: Lane 1, 1 kb DNA ladder; Lane 2 and 4, WB gDNA; Lane 3, IFT121-mNG gDNA; Lane 4, IFT140-mNG gDNA. (D) PCR analysis of IFT complex B-related genes: Lane 1, 1 kb DNA ladder; Lane 2, 4, 6, 8, and 10, WB gDNA; Lane 3, IFT20-mNG gDNA; Lane 5, IFT46-mNG gDNA; Lane 7, IFT52-mNG gDNA; Lane9, IFT81-mNG gDNA; Lane 11, IFT88-mNG gDNA. (E) PCR analysis of motor protein genes: Lane 1, 1Kb DNA ladder; Lane 2 and 4, WB gDNA; Lane 3, Kin-2b-mNG gDNA; Lane 5, Kin-13-mNG gDNA. Arrows indicate PCR products expected to be amplified from the mNG-tagged genes. Asterisks indicate a nonspecific PCR product.
In case of 5 components for IFT complex B, PCR on all of the IFT-mNG candidates resulted in PCR products at the expected sizes (Fig. 1D) whereas the same PCR did not produce any DNA using Giardia WB gDNA as a template. Additional nonspecific DNA fragments were found in PCR with IFT52-mNG candidate.
Integration of 2 mNG-tagged kinesin (kinesin-13 and kinesin-2b) genes into Giardia chromosome was also detected by PCR (Fig. 1E).
Expression of mNG-tagged IFT proteins were examined in the constructed transfectants under a fluorescence microscope. Giardia trophozoite has a morphology with polarity, i.e., it has stumpy anterior end, slender posterior end, concave front, and convex back with bilateral symmetry (Fig. 2A). Two nuclei and an adhesive disc are located closer to anterior than posterior ends. Flagella were composed of 2 parts, cytoplasmic and membranous portions. All 4 pairs of flagella have a direction from anterior pointing to the posterior end of trophozoites. Structures of 4 pairs of flagella are distinct in that they are extruded from the distinct locations of the Giardia cells. The starting points of membranous flagella extended from cytoplasmic flagella are called flagella pore (FP) as indicated in Fig. 2A. In case of cells expressing mNG-tagged IFT121 and IFT140, components of the IFT complex A, both of them were present in cytoplasmic part (axonemes) of ventral and posterolateral flagella and also found in the FPs for membranous flagella of anterior, ventral, and posterolateral pairs (Fig. 2B). For 5 members of IFT complex B, all of them were present in axonemes of posterolateral flagella (Fig. 2C). On the other hand, they were different with respect to localization at FPs. IFT20 were found at FPs of anterior, ventral, and caudal flagella whereas IFT46 and IFT81 were present at FPs of anterior and ventral flagella. The rest 2 IFTs, IFT52 and IFT88 were detected only at the FPs for anterior flagella. Lastly, localization of 2 motor proteins for IFT, were also examined in Giardia cells expressing kinesin-13-mNG or kinesin-2b-mNG (Fig. 2D). Fluorescence was only present in median bodies in the cells carrying the kinesin-13-mNG. Cells expressing kinesin-2b-mNG demonstrated strong fluorescence at the cytoplasmic axonemes for all flagella.
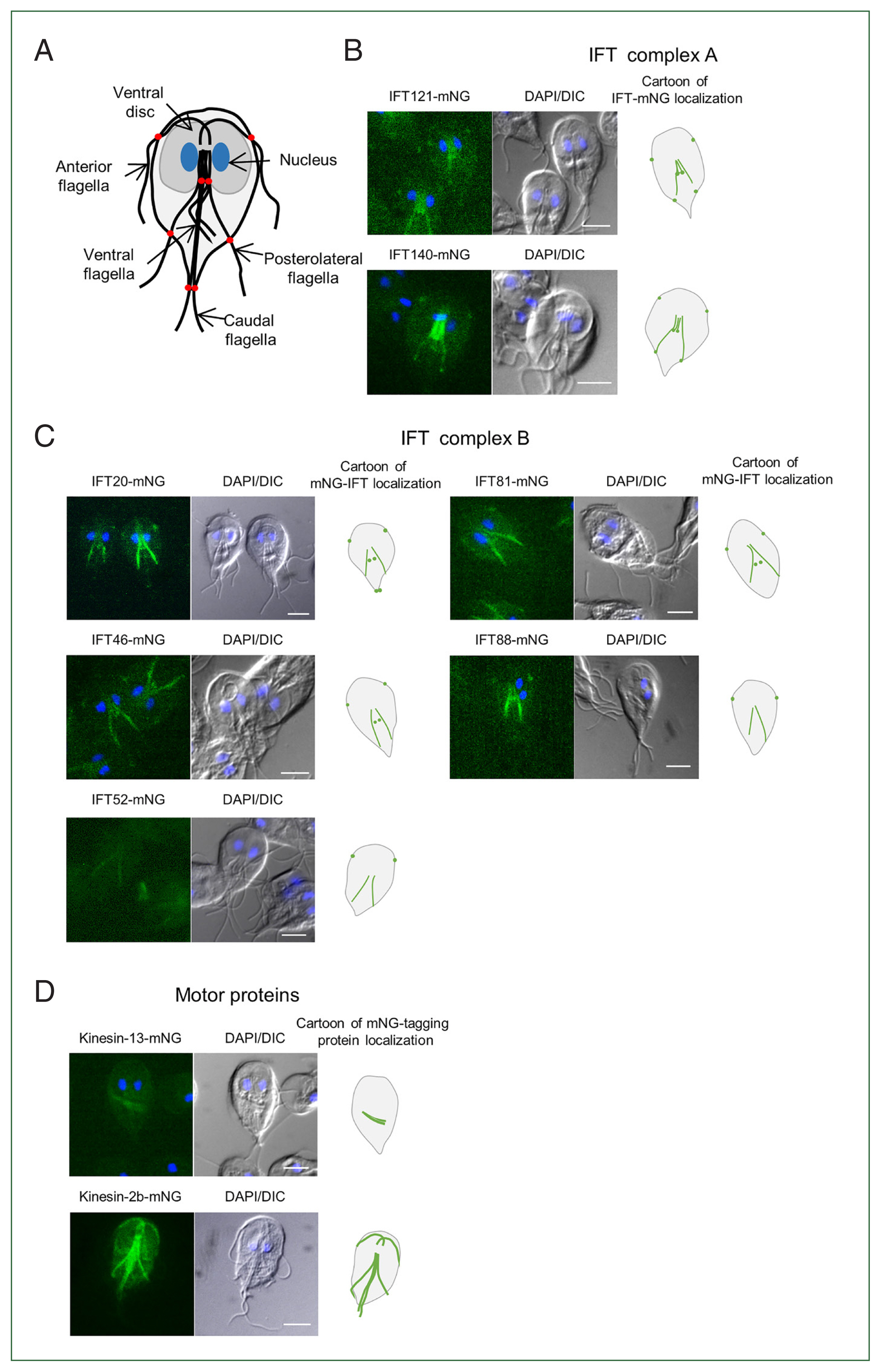
Localization of mNeonGreen (mNG)-tagged intraflagellar transport (IFT) complex proteins in Giardia trophozoites. (A) Illustration of Giardia trophozoite. The cell has 2 nuclei (blue), an adhesive disc (dark gray), and 4 pairs of flagella; anterior, posterolateral, ventral, and caudal. The red dots in the cartoon indicate the flagellar pores. The localization of IFT complex proteins was observed on the fixed cells using fluorescence microscopy. (B) Localization of the IFT complex A-components, IFT121- and IFT140-mNG proteins. (C) Localization of the IFT complex B-components, IFT20-, 46-, 52-, 81-, and 88-mNG proteins. (D) Localization of the motor proteins, kinesin-13 and kinesin-2b. All scale bars are 5 μm.
Effect of dCas9-mediated IFT88 KD in flagella formation
Among the IFT components we had been investigated, we paid more attention to the IFT complex B, which is known as an anterograde IFT train for flagella formation. IFT88 were chosen for further studies because its expression was more distinct in fluorescent images of IFT88-mNG expressing Giardia cells (Fig. 2C). Transgenic Giardia cells with decreased level of IFT88 expression were constructed using dCas9-mediated KD system as described [10]. We first established Giardia expressing dCas9 along with 1 of the 3 gRNAs which were designed to interfere the transcription of ift88 gene at +33, +258, or +455 relative to the start codon of IFT88 (Supplementary Table S2). As a control, a random oligonucleotide sequence was cloned into dCas9-expressing plasmid and transfected into Giardia, and the resulting transfectants were monitored for a transcriptional level of ift88 mRNA. Two of the gRNAs annealed to +33 and +455 did not affect the production of ift88 transcript, whereas the +258 gRNAs caused a decrease in ift88 mRNA as determined by quantitative real-time PCR (data not shown). Especially the gRNA at +258 decreased the transcription to 65.6% of the cells with control gRNA, and we used this transfectant as the IFT88 KD strain (Fig. 3A).
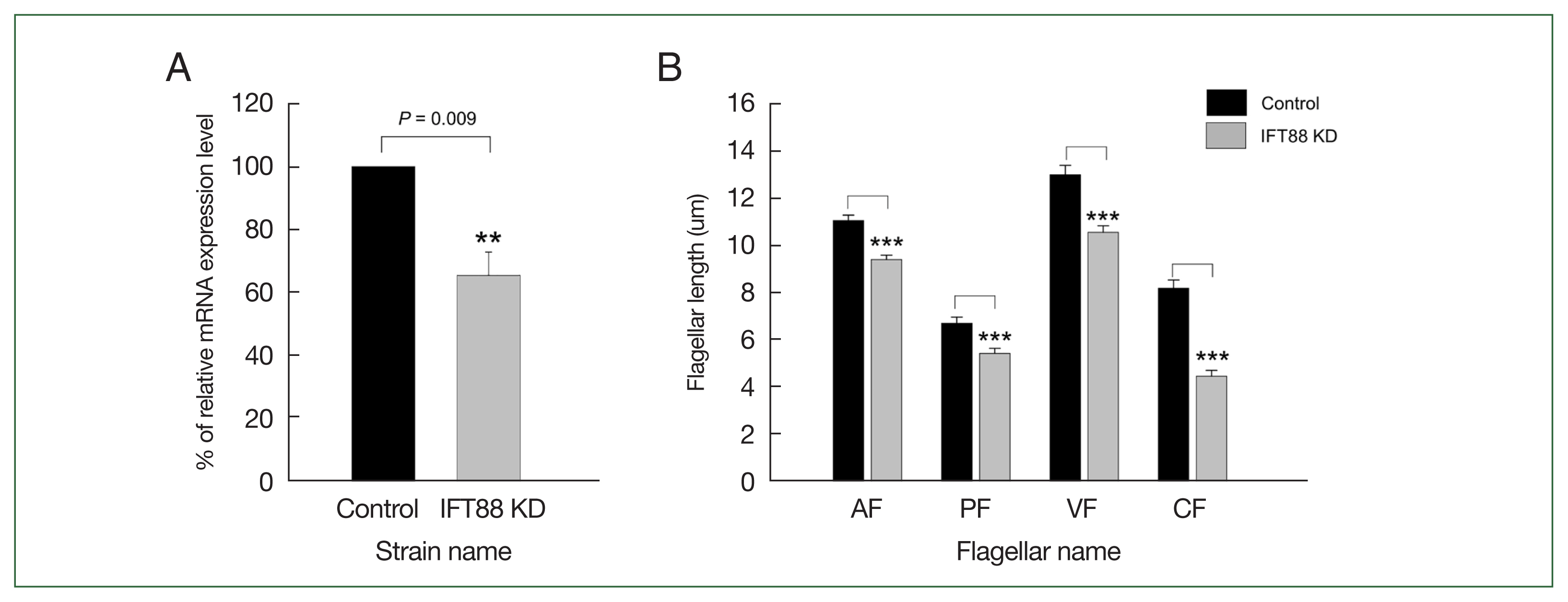
CRISPRi-mediated IFT88 knockdown (KD) altered Giardia flagellar length. (A) IFT88 KD was confirmed by quantitative real-time PCR. The gene expression of ift88 in the IFT88 KD cells (gray bar) was compared with the nonspecific guide RNA-expressing cells (closed bar). IFT88 expression was normalized to that of actin expression in the same cells. (B) Effect of IFT88 KD on lengths of 4 pairs of flagella, designated anterior (AF), posterolateral (PF), ventral (VF), and caudal (CF). Both IFT88 KD (gray bars) and control cells (closed bars) were stained with Giemsa solution and scored for flagellar length. At least 35 cells were analyzed for each strain. Data are presented as the mean of 3 independent experiments. In all panels, error bars represent 95% confidence intervals, and significance was assessed using Student t-test. **P<0.01 and ***P<0.0001.
In addition, we examined whether the IFT KD affected the flagella formation (35 cells per each condition, and 3 independent experiments) by quantitatively measuring the length of the membrane-bound portion of all 4 pairs of flagella (Fig. 3B). The control cells, i.e., Giardia cells with the control gRNA, demonstrated the membrane-bound ventral flagella as the longest (13.0 μm) whereas the posterolateral flagella had the shortest membrane-bound region (6.7 μm). The lengths of the anterior and caudal membrane flagella were 11.1 and 8.2 μm, respectively. In IFT88 KD Giardia trophozoites, the lengths of all 4 kinds of flagella found to be decreased with statistical significance (ventral=10.6 μm, posterolateral=5.5 μm, anterior=9.2 μm, and caudal=4.5 μm).
Effect of dCas9-mediated kinesin-2b in flagella formation
IFT88 is 1 of IFT complex B which plays a role in anterograde transport system of flagella formation. Therefore, we investigated the role of a motor protein, kinesin-2 for anterograde IFT train in the subsequent experiment. We designed three gRNAs targeting at +21, +141, and +271 within the open reading frame of kinesin-2b, 1 of 3 subunits of heterotrimeric kinesin-2 (Supplementary Table S2). Expression of kinesin-2b +271 in dCas9-expressing Giardia trophozoites decreased kinesin-2b mRNA (Fig. 4A), whereas kinesin-2b +21 and +141 gRNAs did not cause any changes in transcription of kinesin-2b mRNA (data not shown).
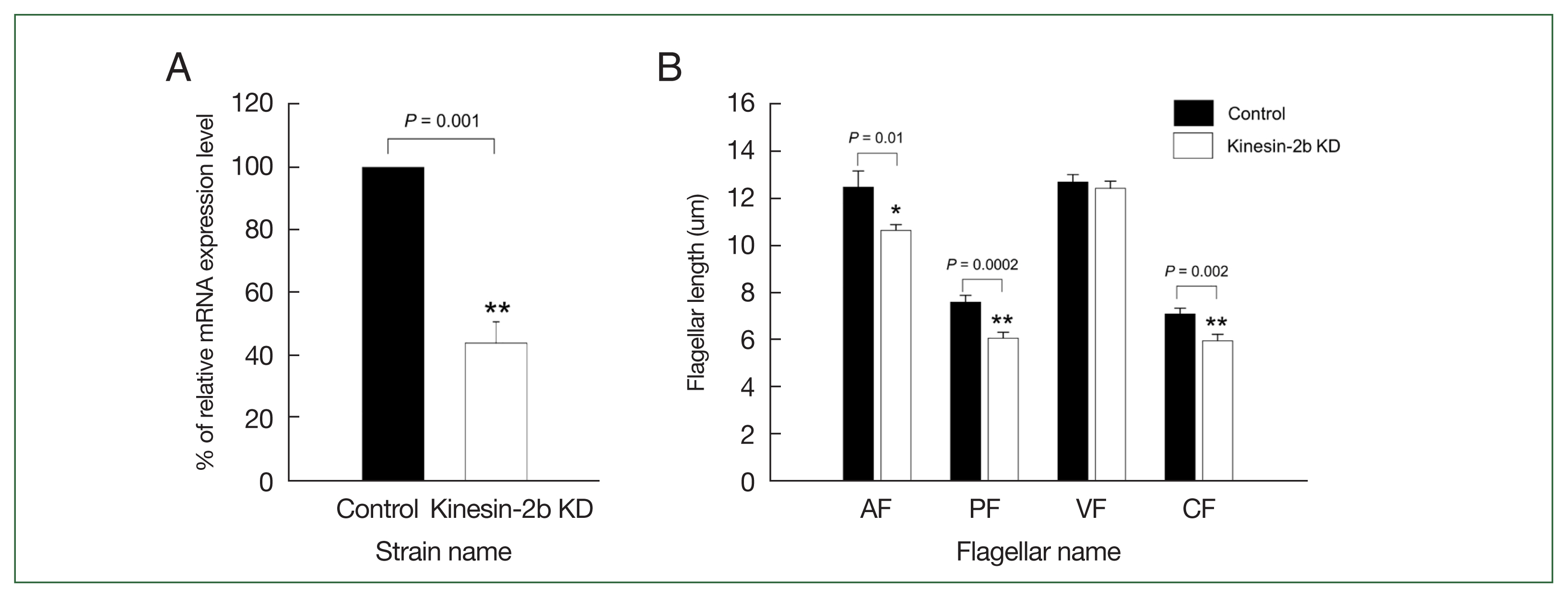
CRISPRi-mediated kinesin-2b knockdown (KD) caused alteration of Giardia flagellar length. (A) Determination of kinesin-2b mRNA level in kinesin-2b KD cells (open bars) and control cells (closed bars) by quantitative real-time PCR. The expression of kinesin-2b transcript was normalized to that of Giardia actin. (B) Effect of kinesin-2b KD on flagellar length of anterior (AF), posterolateral (PF), ventral (VF), and caudal flagella (CF). Control (closed bars) and kinesin-2b KD cells (open bars) were stained with Giemsa solution. At least 35 cells were analyzed for each group. Data are presented as the mean of 3 independent experiments. Student t-test, **P<0.01.
Giardia trophozoites carrying pdCas9-Kin2b-g271 demonstrated decreased level of kinesin-2b mRNA to 44.3% of the control cells, and we used this strain as kinesin-2b KD. Therefore, these cells were scored for flagella length as described above (Fig. 4B). Kinesin-2b KD cells showed shortened lengths of three pairs of flagella, anterior (from 12.5 μm of the control cells to 10.7 μm), posterolateral (from 7.6 μm of the control to 6.1 μm), and caudal flagella (from 7.1 μm of the control cells to 6.0 μm), whereas the length of ventral flagella was not different from that of control cells.
Presence of IFT88 at axonemes and 8 FPs in live Giardia cells
When we observed the fixed cells expressing IFT88-mNG, IFT88 was detected in axoneme of posterolateral flagella and FPs of anterior flagella (Fig. 2C). Giardia cells expressing IFT88-mNG was examined via live-cell imaging in order to confirm the result on the fixed cells and to improve the sensitivity of the imagining (Fig. 5A). Live imaging of these cells up to 140 sec clearly demonstrated localization of fluorescence in axonemes for ventral and posterolateral flagella (Supplementary Video S1). In addition, presence of IFT88-mNG was detected in the FPs for all 4 pairs of flagella. Fluorescence images of Giardia cells expressing IFT88-mNG at 6 specific time points (0, 31, 63, 93, 121, and 140 sec) of the live imaging clearly demonstrate the localization of IFT88 in the FPs and axonemes (Fig. 5B).
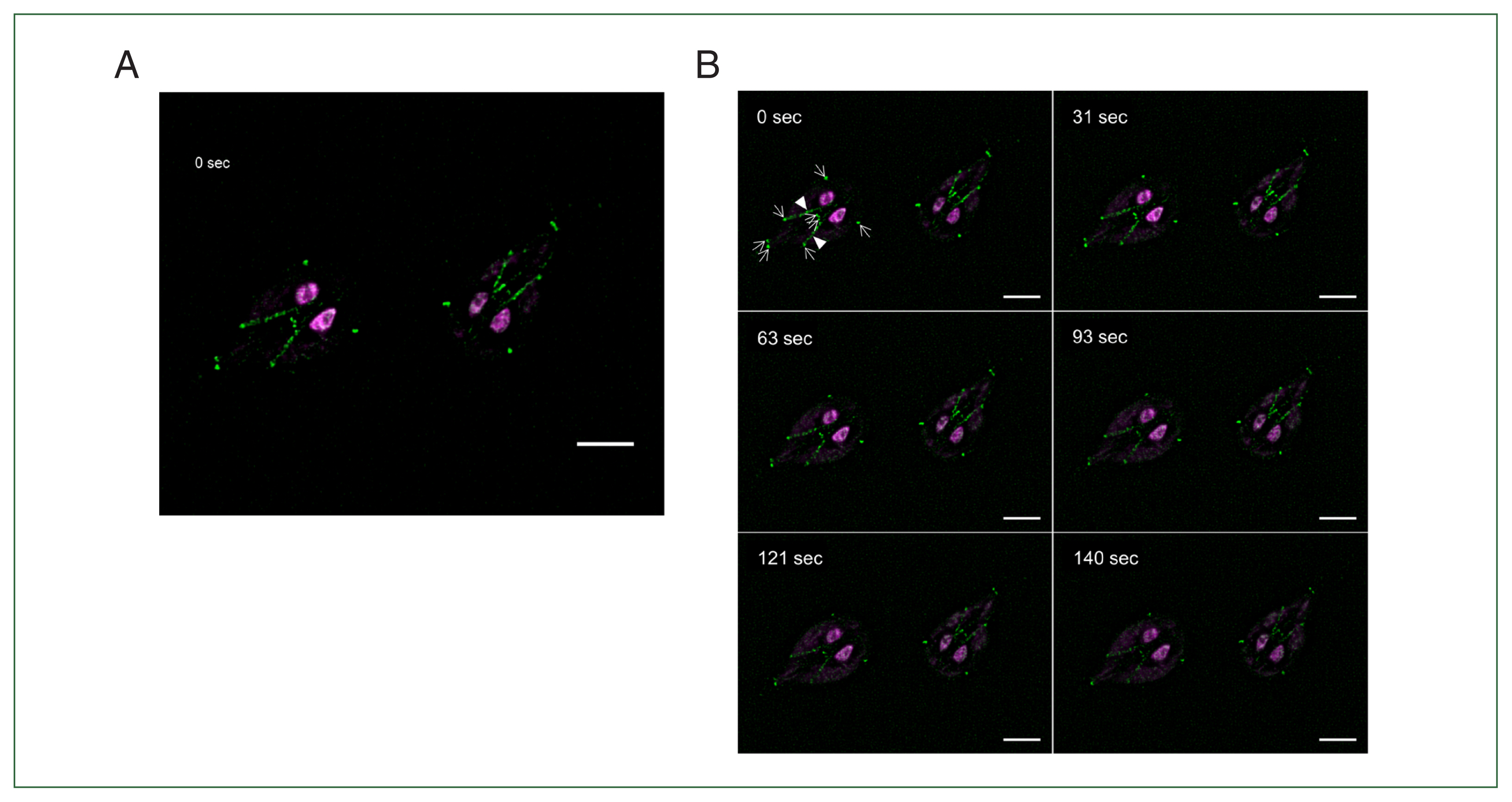
IFT88 accumulates in flagella pore (FP) region. (A) Live-cell imaging of Giardia trophozoites expressing IFT88-mNeonGreen. Live-cell imaging was recorded for 140 sec (Supplementary Video S1). Video was recorded at 1 frame per second and played at 20×increased speeds. Time is indicated in the upper left corner. (B) Time-series images were presented at approximately 30 sec intervals (0, 31, 63, 93, 121, and 140 sec). Time is indicated in the upper left corner. IFT88-mNeonGreen was observed mainly in posterolateral and ventral cytoplasmic axonemal regions and accumulated in 8 FP regions of the Giardia cells. Arrows indicate 8 FPs, and arrowheads indicate posterolateral cytoplasmic axonemes. Scale bar=5 μm.
Discussion
This study characterized the role of IFT components in flagella formation in the flagellate protozoan Giardia lamblia. Cilia or flagella biogenesis involves IFT, which was first discovered in Chlamydomonas [20–22]. Since its first discovery, IFT has been founded to be existed in most eukaryotic cilia and flagella [7]. The IFT particles consist of the retrograde IFT complex A and the anterograde IFT complex B. The IFT complex B includes at least 16 members, is linked to a kinesin-2 motor and participates in anterograde transport from the cell body to the flagellar tip during flagellar assembly. The IFT complex A contains at least 6 members, is connected to a dynein motor and is responsible for retrograde transport from the flagellar tip to the cell body during flagellar recycling [23]. Based on GiardiaDB, Giardia has three IFT complex A components (IFT121, 122, and 140) and 12 IFT complex B components (IFT20, 38, 46, 52, 54, 56, 57, 74/72, 81, 88, 172, and 180). In this study, we focused on whether the anterograde IFT complex B components and kinesin-2 motor protein are involved in Giardia flagellar assembly.
Giardia trophozoite has 4 pairs of bilateral and symmetrical flagella (anterior, posterolateral, ventral, and caudal), and each pair of flagella has a different length. Each of the 8 flagella has a long cytoplasmic axoneme before exiting the cell body as a membrane-bound flagellum (Fig. 2A) [11–13,24]. In this study, we generated cells that expressed IFT and motor protein with a C-terminal mNG tag at their genomic locus to ensure that overexpression of these proteins would not interfere with their function (Fig. 1A, B). All constructs tagged with mNG were integrated and maintained within their genomic loci without affecting their expression (Figs. 1, 2). We observed the localization of 2 IFT complex A proteins (IFT121 and 140) and 5 IFT complex B proteins (IFT20, 46, 52, 81, and 88) in fixed cells, and confirmed to be present in the FPs and axonemes (Fig. 2B–D). Previous studied by McInally et al. showed that IFT complex A proteins (IFT120, 121, and 140) and some IFT complex B proteins (IFT30, 54, 56, 57, 74, 80, 81, 88, and 172) were localized to FPs and axonemes [9]. Our results are consistent with those of previous studies. In addition, our results provide evidence that IFT20, 46, and 52 are the IFT complex B proteins in Giardia (Fig. 2C).
In this study, we established a live-cell imaging method using IFT88-mNG expressing cell under a high-resolution microscope to observe the movement of IFT particles. As expected, IFT88-mNG protein was observed to move between cytoplasmic axonemes of posterolateral and ventral flagella and the FPs (Fig. 5). In other organisms, IFT trains assemble and accumulate at the TZ. The TZ is a structurally and functionally distinct region where anterograde IFT proteins are loaded and transported to the distal flagellar tip by the heterotrimeric kinesin-2 complex [25,26]. Interestingly, Giardia lacks a canonical TZ and TZ-related proteins [7,8]. Therefore, McInally et al. suggested that Giardia 8 FP complexes are functionally analogous to the TZ in other organisms because IFT components and kinesin-2 motor protein were concentrated between the cytoplasmic and membrane-bound axoneme compartment [9]. Although we did not observe the translocation of the IFT88 protein from the FPs to the flagellar tips, we confirmed that this protein accumulated at all FPs (Fig. 5). We also observed that other tagged IFT complex proteins in this study were also accumulated at FPs (Fig. 2). Thus, our results seem to support the model that Giardia FPs are the sites of IFT injection into membrane-bound axonemes.
Interestingly, in fixed cell, IFT88-mNG protein was observed in posterolateral axonemes and anterior FP (Fig. 2C), whereas a live-cell imaging revealed that this protein was present in posterolateral and ventral axonemes and all 4 pairs of FPs (Fig. 5). These results showed that high-resolution microscope is essential for observing IFT proteins expressed in small amounts.
A method using dCas9 was used to generate KD cells for IFT proteins in Giardia instead of morpholinos [9]. This method has the advantage that the inhibition of gene expression can be maintained in a more stable pattern. In cells with KD of IFT88 by dCas9, all flagella were shortened (Fig. 3). Recently, IFT88 protein was reported to be required for the formation of astral MTs and mitotic spindles during the cell cycle in mice and zebrafish [27,28]. During Giardia cell division, 4 mature flagella are structurally inherited (anterior and caudal) and 4 new flagella (posterolateral and ventral) are assembled de novo in each daughter cell [15,29]. Additionally, IFT88 associated with IFT46 and IFT52 to form a trimeric complex as core assembly of IFT complex B in Chlamydomonas [30,31]. Therefore, further studies are needed to determine whether giardial IFT88, 46, and 52 form a complex for flagella formation. The kinesin-2 protein is composed of a heterotrimeric complex of kinesin-2a, kinesin-2b, and kinesin-associated protein and move the anterograde IFT complex B components to the flagellar tips [4]. Similar to other eukaryotes, kinesin-2a regulate flagellar assembly in Giardia, as the overexpression of dominant negative or CRISPRi-mediated KD result in dramatic flagellar length defects [9,32]. This study demonstrated that KD of kinesin-2b resulted in defects in three flagella pairs, as expected (Fig. 4).
The characterization of the ultrastructure and the composition of the FP complex in Giardia will be important in determining how this complex mediates compartmentalization and regulates IFT injection. Recent studies in Giardia have shown that cell-cycle related kinases, such as polo-like kinase (PLK), cyclin-dependent kinase 1 (CDK1), and Nek kinase (NEK), are involved in flagella assembly. Depletion of PLK and CDK1 resulted in long flagella, while depletion of NEK8445 resulted in shorten flagella [33–36]. In addition, PLK and CDK1 have been observed to be localized to the FPs [33,35]. Although NEK8445 was not observed at the FPs, malformation of the caudal FPs was observed in NKE8445-depleted cells [36]. These results suggest that kinases could regulate not only IFT proteins and motor proteins but also FP-related proteins in the FPs of Giardia lacking TZ. Therefore, further studies should be performed to observe the modification of IFT proteins and motor proteins by these signaling components.
In conclusion, we established a system for observing flagellar formation in G. lamblia using mNG-tagged IFTs and investigated their localization. Using CRISPRi-mediated KD experiments, IFT88, a putative component of IFT complex B, and its cognate motor protein kinesin-2b were shown to be involved in the flagella assembly of Giardia lamblia.
Notes
Author contributions
Conceptualization: Kim J, Park SJ
Data curation: Kim J, Park SJ
Formal analysis: Kim J
Funding acquisition: Park SJ
Investigation: Yeo HR, Shin MY, Kim J
Methodology: Yeo HR, Shin MY, Kim J
Project administration: Kim J, Park SJ
Supervision: Kim J, Park SJ
Validation: Park SJ
Writing – original draft: Kim J, Park SJ
Conflict of interest
The authors declare no conflict of interest related to this study.
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Korea government (MSIT) (NRF-2023R1A2C1002475) and by a faculty research grant of Yonsei University College of Medicine (6-2022-0043).
Supplementary Information
Supplementary material is available with this article at https//doi.org/10.3347/PHD.24064.
