Abstract
The mainstay therapy against leishmaniasis is still pentavalent antimonial drugs; however, the rate of antimony resistance is increasing in endemic regions such as Iran. Understanding the molecular basis of resistance to antimonials could be helpful to improve treatment strategies. This study aimed to recognize genes involved in antimony resistance of Leishmania tropica field isolates. Sensitive and resistant L. tropica parasites were isolated from anthroponotic cutaneous leishmaniasis patients and drug susceptibility of parasites to meglumine antimoniate (Glucantime®) was confirmed using in vitro assay. Then, complementary DNA-amplified fragment length polymorphism (cDNA-AFLP) and real-time reverse transcriptase-PCR (RT-PCR) approaches were utilized on mRNAs from resistant and sensitive L. tropica isolates. We identified 2 known genes, ubiquitin implicated in protein degradation and amino acid permease (AAP3) involved in arginine uptake. Also, we identified 1 gene encoding hypothetical protein. Real-time RT-PCR revealed a significant upregulation of ubiquitin (2.54-fold), and AAP3 (2.86-fold) (P<0.05) in a resistant isolate compared to a sensitive one. Our results suggest that overexpression of ubiquitin and AAP3 could potentially implicated in natural antimony resistance.
-
Key words: Leishmania tropica, antimony resistance, cDNA-AFLP, real-time RT-PCR, ubiquitin, amino acid permease
INTRODUCTION
Leishmaniasis is one of the tropical diseases caused by infection with the dimorphic protozoan parasites
Leishmania. The clinical manifestations vary from self-healing cutaneous lesion to fatal visceral disease [
1]. Cutaneous leishmaniasis (CL) is the most common form of the disease which has been reported from 77 countries with incidence of 1-1.5 million annual new cases [
2]. CL is an important public health problem in different parts of Iran and 13-25% of reported CL in some parts of Iran are anthroponotic cutaneous leishmaniasis (ACL) caused by
Leishmania tropica [
3,
4]. Chemotherapy still remains the sole choice for treatment and antimonial compounds such as meglumine antimoniate (Glucantime®) have been in use for more than 70 years [
5]. Unfortunately, in recent years antimonial therapy has been imperiled due to an increasing rate of treatment failure attributed to antimony resistant parasites in some endemic foci, especially in India where 37-70% of visceral leishmaniasis patients failed to respond after antimonial therapy [
6]. Failure of antimonial treatment was also reported in 10-12% of ACL patients in Iran which was shown to be due to drug resistance [
7]. Most of the knowledge regarding antimony resistance process has derived from laboratory generated parasites suggesting presence of different mechanisms such as decreased capacity of converting SbV to SbIII, reduced drug uptake, and elevated drug efflux [
8]. In laboratory generated parasites, elevated levels of trypanothione [
9] and downregulation of aquaglyceroporin (AQP1) which uptake trivalent metalloid [
10] have been demonstrated. Additionally, overexpression of ATP-binding cassette (ABC) protein MRPA which sequestered Sb-thiol complexes has been related to the drug resistance [
11].
Recent studies on resistant field isolates have suggested that in addition to the conventional mechanisms other factors such as apoptosis might be implicated in natural antimony resistance [
12,
13]. Considering that, more insight into the mechanisms responsible for natural antimony resistance could improve current treatment strategies.
Several techniques such as RT-PCR, microarrays, and proteomics are currently used to explore molecular markers of antimony resistance [
14,
15]. cDNA-amplified fragment length polymorphism (cDNA-AFLP) is one of the most sensitive technologies for detection of differentially expressed gene which does not require any prior information of gene sequences [
16,
17]. In this study, cDNA-AFLP was used to identify the potential biomarkers of antimonial drug resistance in
L. tropica field isolates.
MATERIALS AND METHODS
Clinical isolates and cell culture
L. tropica field strains used in the present study included meglumine antimoniate sensitive (MHOM/IR/10/Mash-175) and resistant (MHOM/IR/10/Mash-827) isolates which were obtained from patients with ACL in Mashhad, north-east of Iran [
7]. Unresponsiveness was defined as patients who regardless of receiving at least 3 full courses of Glucantime® still showed active lesions [
7]. The Institutional Ethical Committee of the School of Public Health, Tehran University of Medical Sciences reviewed and approved the study protocol.
The parasites within 6 subcultures after the isolation from the patients were used. Cryopreserved parasites were recovered from liquid nitrogen and cultured in RPMI 1640 medium (Gibco/BRL, Rockville, Maryland, USA) supplemented with 10% fetal bovine serum (FBS; Gibco/BRL), 100 U/ml penicillin, 100 µg/ml streptomycin (Gibco/BRL), in the absence of antimonial drug pressure, and incubated at 26℃.
In vitro drug sensitivity assay
The drug susceptibility of the field isolates in monocyte-macrophage mouse cell-line (J774A.1) was analyzed in triplicate as describe previously with slight modifications [
7]. Briefly, J774A.1 (5×10
4 cells/well) were grown in RPMI with 10% FBS in Labtek 8-well chamber slides (Nunc, New York, USA) and incubated at 37℃ for 24 hr to allow cell attachment, then the cells were infected with stationary phase promastigotes at a parasite to macrophage ratio of 10:1. After 4 hr incubation at 37℃, cells were washed for 3 times and incubated for 72 hr in the presence of serial dilution of Glucantime® (Sanofi-Aventis, Paris, France). Pentavalent antimony concentrations used for the sensitive isolate were 2, 4, 6, 8, 10, and 12 µg/ml, and the for resistant isolate were 35, 40, 45, 50, 55, 60, 65, and 70 µg/ml (the doses were obtained based on previous screening test). The fresh drug was added, and slides were incubated for an additional 72 hr, and then stained using Giemsa. The number of amastigotes in 100 randomly chosen macrophages was counted. The inhibitory concentration 50% (IC
50) was calculated using the values for the number of amastigotes/macrophage. IC
50 was defined as the effective dose of Glucantime® that reduces the survival of
L. tropica by 50%.
Total RNA extraction was carried out using Tripure isolation reagent (Roch, Mannheim, Germany) according to the manufacture's instructions with minor modifications. Briefly, 1×108 log-phase promastigotes of sensitive and resistant isolates were harvested and lysed using Tripure reagent. The lysate was mixed with chloroform and vortexed vigorously, and then centrifuged at 12,000 g for 15 min at 4℃. Afterwards, the upper colorless phase was precipitated by adding isopropanol and washed with 75% ethanol. Finally, the precipitated RNA was dried at room temperature and dissolved in RNase free water. The quantity and quality of RNA were analyzed using nanodrop (ND-1000, Thermo Scientific Fisher, Waltham, Massachusetts, USA) and gel electrophoresis, respectively. To avoid any genomic contamination, RNA was treated with DNase (Qiagen, Hilden, Germany) according to the manufacture's protocols.
cDNA-AFLP
The cDNA-AFLP assay was conducted as describe previously with some modifications [
18]. Single strand cDNA (sscDNA) was synthesized using 10 µg RNA, 20 pmol/µl oligo-dT (Fermentas, Burlington, Canada), 20 pmol/µl random hexamer (Fermentas), 10 mM of dNTP mix (Fermentas) incubated at 65℃ for 10 min, followed by addition of 20 U RNase inhibitor (Fermentas), 4 µL of 5× reverse transcriptase (RT) buffer containing Tris-HCl (pH 8.3) (Fermentas), and 200 U RevertAid premium RT (Fermentas). It was then incubated at 25℃ for 10 min followed by incubation at 50℃ for 60 min. The integrity of cDNA was checked using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) primers as the housekeeping gene (
Table 1). The PCR condition was an initial denaturing step of 94℃ for 5 min and 30 repetitions of denaturation at 94℃ for 30 sec, annealing at 60℃ for 30 sec, and extension at 72℃ for 45 sec with a final extension of 72℃ for 7 min.
Double strand cDNA (dscDNA) synthesis was performed using 40 U DNA Polymerase I (Fermentas), 1.5 U Ribonuclease H (Roche, Mannheim, Germany), and 10 mM of dNTP mix at 16℃ for 2 hr, followed by purification with High Pure PCR Clean up Micro Kit (Roche). Double restriction digestion was carried out with 5 U MboI and 5 U EcoRI (Fermentas) on 5 µg purified dscDNA at 37℃ for 2 hr.
Digested dscDNA fragments were ligated to AFLP adaptors (
Table 1), 8 µg AD Mbo1, AD EcoR1, 4 µg ad Mbo1, and ad EcoR1 in a final volume of 60 µl. Briefly, it was conducted in the following conditions: 1 min at 55℃, decreasing to 10℃ over 45 min (i.e., 1℃ per 1 min) and then 6 U T4 DNA Ligase (Roche, Mannheim, Germany) was added and incubated at 4℃ overnight. After purification with High Pure PCR Clean up Micro Kit (Roche), pre-amplification with pre-amp primers (
Table 1) was performed according to the following conditions: 30 cycles of 94℃, 30 sec, 64℃, 30 sec, and 72℃, 45 sec, and final extension at 72℃ for 7 min.
The sensitive amplification was done using various dilutions of pre-amplified products to get the optimal conditions. PCR amplification was carried out as described above using sensitive primers containing adaptor sequences plus 1 nucleotide at 3' end (
Table 1).
The products resulting from sensitive amplifications were resolved on 8% non-denaturating PAGE and stained using silver nitrate. Finally the gels were wrapped and scanned, and then checked for the presence of differentially expressed transcription-derived fragments (TDFs).
Isolation, cloning, and sequencing of TDFs
Selected bands were extracted from the polyacrylamide gels. The eluted DNA was re-amplified with selective primers (
Table 1) from resultant profile. After checking PCR products on 1.5% agarose gel, they were cloned into a pGEM-T Vector System I (Promega, Fitchburg, Wisconsin, USA). To identify the isolated TDFs, recombinant plasmid containing an unknown DNA was subjected to sequencing using universal primers (T7 promotor and SP6) (Bioneer, Seoul, South Korea). Homology searches were performed in non-redundant nucleic and protein databases BLAST (
http://www.ncbi.nlm.nih.gov/BLAST).
In order to confirm the differences in expression patterns of TDF-derived genes between resistant and sensitive
L. tropica isolates, real-time RT-PCR was conducted. Target gene primers were designed by Primer 3 software version 0.4.0 (
http://frodo.wi.mit.edu) according to the identified genes (
Table 1). The cDNA was synthesized from 1 µg of total RNA using QuantiTect® Reverse Transcription Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. RT-PCR was performed in triplicate with 20 µl volumes using SYBR® Premix Ex Taq™ II (Takara, Tokyo, Japan) in a CFX96™ Real-Time System (C1000™ Thermal Cycler) (BioRad, Hercules, California, USA). The PCR condition was as follows: activation at 95℃ for 30 sec, amplification at 95℃ for 5 sec, 60℃ for 30 sec for 40 cycles. The relative value of the expression level of each gene was calculated by comparing the cycle threshold (
CTs) of the target genes with that of the housekeeping gene (GAPDH) using the 2
-ΔΔct method [
19].
In vitro drug susceptibility assay was presented as IC50 which was determined using the linear regression analysis. Each experiment was conducted at least thrice, and the results are expressed as the mean±standard error of the mean (SEM). The significance of differences was determined using the Student's t-test. The level of statistical significance was P<0.05.
RESULTS
Drug susceptibility of Leishmania tropica field isolates
In vitro drug sensitivity of L. tropica to Glucantime® was determined in J774-1 cells. The results showed that IC50 values of Sb (V) for resistant isolate was 50.0±3.5 µg/ml, whereas for sensitive isolate was 6.0±1.5 µg/ml. The IC50 of resistant isolate was about 8-fold higher than the sensitive strain.
The cDNA-AFLP analysis of clinical isolates
To identify differentially-expressed transcripts putatively associated with natural antimony resistance, a cDNA-AFLP approach was utilized on resistant and sensitive
L. tropica field isolates. Total RNA was extracted and the sscDNA was synthesized from each sample. The integrity of cDNA was confirmed by specific primers of the housekeeping gene (GAPDH) (
Fig. 1).
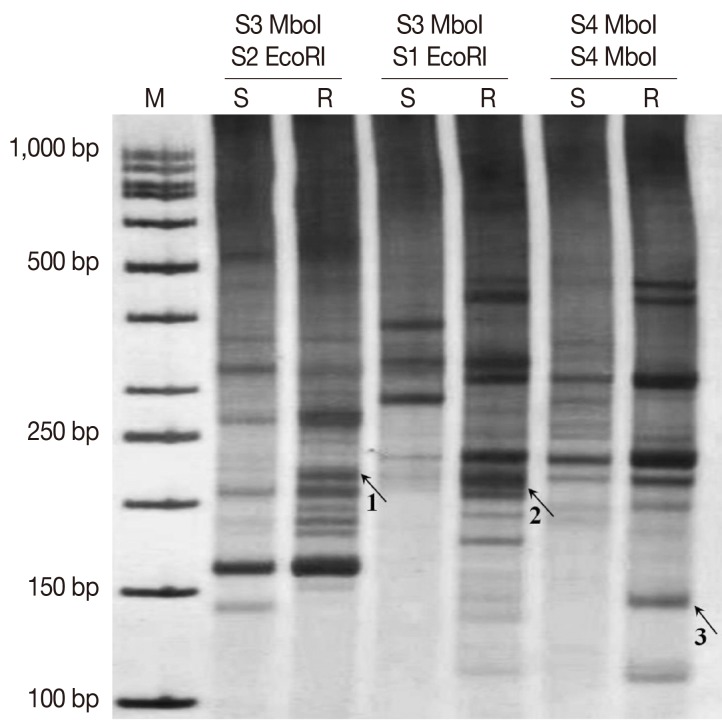
The cDNA AFLP results revealed differential expressions of TDFs on 8% non-denaturating PAGE by silver staining using different primer combinations (
Fig. 2). Differentially-expressed bands were extracted, re-amplified, and cloned into a TA-cloning system. After then, the recombinant plasmids were sequenced. The sequencing results were searched in non-redundant nucleic and protein databases to find homologous sequences and gene identity.
Blast analysis revealed that 1 clone highly matched with ubiquitine implicated in protein degradation and another clone highly homologous to amino acid permease (AAP3) involved in arginine uptake. Moreover, a clone was found to be similar to a gene which coded a hypothetical protein. The details of sequencing results of identified genes are presented in
Table 2.
In order to validate the cDNA-AFLP results, the mRNA expressions of ubiquitin and AAP3 in resistant and sensitive
L. tropica field isolates were analyzed using the real-time RT-PCR.
Fig. 3 shows a significant upregulation of ubiquitin (2.54-fold) and AAP3 (2.86-fold) in the resistant isolate compared to the sensitive one (
P<0.05).
DISCUSSION
Emergence of antimony drug resistance has eroded treatment and control of leishmaniasis in endemic regions such as Iran [
6,
7]. The molecular mechanisms operating in natural antimony resistance are not well known. Different approaches are currently used to elucidate potential factors implicated in this phenomena. In the current study, we utilized cDNA-AFLP to identify genes potentially involved in drug resistance of
L. tropica field isolates. We found 1 unknown gene and also 2 known genes including ubiquitin and AAP3 genes. Furthermore, differential expression of known genes was confirmed by real time RT-PCR. Our findings showed a significant upregulation of ubiquitine and AAP3 mRNA expression in the resistant isolate compared to the sensitive one.
Ubiquitin belongs to the class of heat-shock proteins which play important roles in various cellular processes such as degradation of defective proteins, endocytosis, transcription, DNA repair, and apoptosis [
20]. One of the well understood function of ubiquitin is protein degradation via ubiquitin-proteasome pathway (UPP) by which it prevents cells from toxic accumulation of abnormal proteins [
21]. Through the UPP, ubiquitin attached to lysine residues of the target proteins leading to degradation of the ubiquitin-tagged protein by the 26S proteasome [
22]. Several studies have revealed that the wild-type ubiquitin is cytoprotective and vital for cell surveillance [
21,
22]. Tsirigotis et al. [
23] studied the sensitivity of mammalian cells expressing mutant ubiqutin to protein-damaging agents such as cadmium suggested that genetic alteration within ubiqutin coding region inhibits protein degradation and sensitizes cells to toxic effects of aberrant proteins. Another study showed that in mutant ubiquitin-expressing cells the viability was decreased when the cells were exposed to heat shock and hydrogen peroxide [
24].
It was demonstrated that metalloid compounds such as arsenic and cadmium induce heat shock and/or oxidative stress responses leading to production of aberrant oxidized proteins [
25,
26]. Additionally, these compounds inhibit ubiquitination and degradation of aberrant proteins which might lead to oxidative stress-induced apoptosis [
27]. It was documented that the elevated expression of ubiqutin in organisms such as
Saccharomyces cerevisiae contributes to resistance to oxidative stress through degradation of oxidatively damaged proteins [
28]. Moreover, another study demonstrated that overexpression of ubiquitin in yeast increased ubiquitin-dependent degradation of damaged proteins and could prevent heath shock toxicity and cell death [
29]. Therefore, our results suggest that the overexpression of ubiquitin in antimony resistant
L. tropica isolate could contribute to degradation of oxidized proteins and defend cells from oxidative stress related to antimonial compounds.
Also, our results identified the upregulation of AAP3 in resistant isolate in comparison to the sensitive one. To date several AAPs have been identified in
Leishmania [
30]. AAP3 is a highly specific arginine transporter that belongs to a large amino acid/auxin permease family (AAAP) which localizes to the surface membrane of
Leishmania [
31]. Arginine is known as a precursor of polyamine biosynthesis [
32] which transferred into
Leishmania from external sources through AAP3 [
31]. This amino acid is hydrolyzed by arginase producing ornitine which participate in the synthesis of polyamine and trypanothione [
32]. The trypanothione is a major reduced thiol of
Leishmania parasites and plays a crucial role in detoxification of antimonial drugs [
9,
33]. Furthermore, the association of increased trypanothione levels with antimony resistance has been observed in
Leishmania isolates [
9,
34]. Haimeur et al. [
9] reported that the upregulation of trypanothione in resistant isolates is correlated with overexpression of enzymes involved in the synthesis of polyamines (ornithine decarboxylase; ODC). Based on the above data, it is concluded that an elevated expression of AAP3 in antimony resistant
L. tropica isolate might contribute to enhance the level of trypanothione and consequently detoxification of antimonial drugs.
In conclusion, for the first time, we identified ubiquitin and AAP3 genes as potential antimony resistance candidate target genes in Iranian strains of L. tropica through cDNA-AFLP approach. The overexpression of aforementioned genes might be implicated in natural antimony resistance and could be helpful to get more insight into the molecular etiology of antimony resistance in Leishmania spp. However, further studies are required to establish the exact mechanisms by which these genes act in the resistance process.
ACKNOWLEDGMENTS
The authors would like to thank Mrs. S. Charehdar, Ms. O. S. Dinehkabodi, Dr. Sh. Farahyar, Mr. R. Roozafzoon, Ms. N. Sadeghipour, Dr. khoshzaban, and Dr. Jabarvand for their kind cooperation. This project was financially supported by the Vice Chancellor for Research, School of Public Health, Tehran University of Medical Sciences, Iran (No: 11826). Elham Kazemi-Rad acknowledges support for a Ph.D. degree from Grant No. 240/2151 by Tehran University of Medical Sciences, Tehran, Iran. The authors state that there is no conflict of interests.
References
- 1. Murray HW, Berman JD, Davies CR, Saravia NG. Advances in leishmaniasis. Lancet 2005;366:1561-1577.
- 2. Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J, Jannin J, Boer Md. the WHOLCT. Leishmaniasis worldwide and global estimates of its incidence. PLoS One 2012;7:e35671.
- 3. Statistics of Cutaneous Leishmanaisis in Iran: Hearing before National Leishmaniasis Committee, Office of Zoonoses, Diseases Management Center, Ministry of Health and Medical Education. 2004.
- 4. Talari SA, Talaei R, Shajari G, Vakili Z, Taghaviardakani A. Childhood cutaneous leishmaniasis: report of 117 cases from Iran. Korean J Parasitol 2006;44:355-360.
- 5. Kedzierski L, Sakthianandeswaren A, Curtis JM, Andrews PC, Junk PC, Kedzierska K. Leishmaniasis: current treatment and prospects for new drugs and vaccines. Curr Med Chem 2009;16:599-614.
- 6. Sundar S. Drug resistance in Indian visceral leishmaniasis. Trop Med Int Health 2001;6:849-854.
- 7. Hadighi R, Mohebali M, Boucher P, Hajjaran H, Khamesipour A, Ouellette M. Unresponsiveness to glucantime treatment in Iranian cutaneous leishmaniasis due to drug-resistant Leishmania tropica parasites. PLoS Med 2006;3:e162.
- 8. Ashutosh , Sundar S, Goyal N. Molecular mechanisms of antimony resistance in Leishmania. J Med Microbiol 2007;56:143-153.
- 9. Haimeur A, Guimond C, Pilote S, Mukhopadhyay R, Rosen BP, Poulin R, Ouellette M. Elevated levels of polyamines and trypanothione resulting from overexpression of the ornithine decarboxylase gene in arsenite resistant Leishmania. Mol Microbiol 1999;34:726-735.
- 10. Marquis N, Gourbal B, Rosen BP, Mukhopadhyay R, Ouellette M. Modulation in aquaglyceroporin AQP1 gene transcript levels in drug resistant Leishmania. Mol Microbiol 2005;57:1690-1699.
- 11. El Fadili K, Messier N, Leprohon P, Roy G, Guimond C, Trudel N, Saravia NG, Papadopoulou B, Légaré D, Ouellette M. Role of the ABC transporter MRPA (PGPA) in antimony resistance in Leishmania infantum axenic and intracellular amastigotes. Antimicrob Agents Chemother 2005;49:1988-1993.
- 12. Vergnes B, Gourbal B, Girard I, Sundar S, Drummelsmith J, Ouellette M. A proteomics screen implicates HSP83 and a small kinetoplastid calpain-related protein in drug resistance in Leishmania donovani clinical field isolates by modulating drug-induced programmed cell death. Mol Cell Proteomics 2007;6:88-101.
- 13. Walker J, Gongora R, Vasquez JJ, Drummelsmith J, Burchmore R, Roy G, Ouellette M, Gomez MA, Saravia NG. Discovery of factors linked to antimony resistance in Leishmania panamensis through differential proteome analysis. Mol Biochem Parasitol 2012;183:166-176.
- 14. Do Monte-Neto RL, Coelho AC, Raymond F, Legare D, Corbeil J, Melo MN, Frezard F, Ouellette M. Gene expression profiling and molecular characterization of antimony resistance in Leishmania amazonensis. PLoS Negl Trop Dis 2011;5:e1167.
- 15. Hajjaran H, Azarian B, Mohebali M, Hadighi R, Assareh A, Vaziri B. Comparative proteomics study on meglumine antimoniate sensitive and resistant Leishmania tropica isolated from Iranian anthroponotic cutaneous leishmaniasis patients. East Mediterr Health J 2012;18:165-171.
- 16. Vuylsteke M, Peleman JD, van Eijk MJ. AFLP-based transcript profiling (cDNA-AFLP) for genome-wide expression analysis. Nat Protoc 2007;2:1399-1413.
- 17. Farahyar S, Zaini F, Kordbacheh P, Rezaie S, Safara M, Raoofian R, Heidari M. Overexpression of aldo-keto-reductase in azole-resistant clinical isolates of Candida glabrata determined by cDNA-AFLP. DARU 2013;21:1-7.
- 18. Saffari M, Dinehkabodi OS, Ghaffari SH, Modarressi MH, Mansouri F, Heidari M. Identification of novel p53 target genes by cDNA AFLP in glioblastoma cells. Cancer Lett 2009;273:316-322.
- 19. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔct method. Methods 2001;25:402-408.
- 20. Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol 2005;6:599-609.
- 21. Sherman MY, Goldberg AL. Cellular defenses against unfolded proteins: A cell biologist thinks about neurodegenerative diseases. Neuron 2001;29:15-32.
- 22. Ciechanover A. Intracellular protein degradation: from a vague idea, through the lysosome and the ubiquitin-proteasome system, and onto human diseases and drug targeting (nobel lecture). Angew Chem Int Ed Engl 2005;44:5944-5967.
- 23. Tsirigotis M, Zhang M, Chiu RK, Wouters BG, Gray DA. Sensitivity of mammalian cells expressing mutant ubiquitin to protein-damaging agents. J Biol Chem 2001;276:46073-46078.
- 24. Fujimuro M, Nishiya T, Nomura Y, Yokosawa H. Involvement of polyubiquitin chains via specific chain linkages in stress response in mammalian cells. Biol Pharm Bull 2005;28:2315-2318.
- 25. Del Razo LM, Quintanilla-Vega B, Brambila-Colombres E, Calderón-Aranda ES, Manno M, Albores A. Stress proteins induced by arsenic. Toxicol Appl Pharmacol 2001;177:132-148.
- 26. Leonard SS, Harris GK, Shi X. Metal-induced oxidative stress and signal transduction. Free Radic Biol Med 2004;37:1921-1942.
- 27. Klemperer N, Pickart C. Arsenite inhibits two steps in the ubiquitin-dependent proteolytic pathway. J Biol Chem 1989;264:19245-19252.
- 28. Cheng L, Watt R, Piper P. Polyubiquitin gene expression contributes to oxidative stress resistance in respiratory yeast (Saccharomyces cerevisiae). Mol Gen Genet 1994;243:358-362.
- 29. Friant S, Meier KD, Riezman H. Increased ubiquitin-dependent degradation can replace the essential requirement for heat shock protein induction. EMBO J 2003;22:3783-3791.
- 30. Akerman M, Shaked-Mishan P, Mazareb S, Volpin H, Zilberstein D. Novel motifs in amino acid permease genes from Leishmania. Biochem Biophys Res Commun 2004;325:353-366.
- 31. Shaked-Mishan P, Suter Grotemeyer M, Yoel Almagor T, Holland N, Zilberstein D, Rentsch D. A novel high affinity arginine transporter from the human parasitic protozoan Leishmania donovani. Mol Microbiol 2006;60:30-38.
- 32. Colotti G, Ilari A. Polyamine metabolism in Leishmania: from arginine to trypanothione. Amino Acids 2011;40:269-285.
- 33. Fairlamb AH, Cerami A. Metabolism and functions of trypanothione in the kinetoplastida. Annu Rev Microbiol 1992;46:695-729.
- 34. Mukhopadhyay R, Dey S, Xu N, Gage D, Lightbody J, Ouellette M, Rosen BP. Trypanothione overproduction and resistance to antimonials and arsenicals in Leishmania. Proc Natl Acad Sci USA 1996;93:10383-10387.
Fig. 1Detection of GAPDH as the housekeeping gene (154 bp) by reverse transcriptase-PCR (RT-PCR) on agarose gel (1.5%). M, 50 bp (base pair) molecular weight marker; S, sensitive; R, resistant; NC, negative control.

Fig. 2Expression patterns of TDFs extracted from cDNA-AFLP PAGE. Sensitive amplification of cDNA-AFLP on a PAGE from 3 different primer combinations of S3
MboI/S2
EcoRI, S3
MboI/S1
EcoRI, and S4
MboI/S4
MboI. Arrows point to differentially expressed TDFs which were isolated with code numbers of 1, 2, and 3 (associated genes which derived from these TDFs mentioned in
Table 2). M, 50 bp molecular weight marker; S, sensitve; R, resistant.

Fig. 3The relative expression levels of ubiquitin and amino acid permease (AAP3) genes in sensitive and resistant L. tropica isolates using real-time RT-PCR. The expression of GAPDH was used to normalize the data. Values given are the mean±SD of 3 different experiments (P<0.05). S, sensitive; R, resistant.

Table 1.Sequences of adaptors and primer used in cDNA-AFLP and real-time RT-PCR
Table 1.
|
Name |
Sequences |
Adaptors and primers |
|
AD EcoR1 |
5´-ACCGACGTCGACTATCCATGAAG-3´ |
Adaptors |
|
ad EcoR1 |
5´-AATTCTTCATGG-3´ |
|
|
AD MboI |
5´-CACTATCCAGACTCTCACCGCA-3´ |
|
|
ad MboI |
5´-GATCTGCGGTGA-3´ |
|
|
Pre EcoR1 |
5´-ACCGACGTCGACTATCCATGAAGAATTC-3´ |
Pre-amplification primers |
|
Pre MboI |
5´-CACTATCCAGACTCTCACCGCAGATC-3´ |
|
|
S1 EcoR1 |
5´-ACCGACGTCGACTATCCATGAAGAATTCC-3´ |
Sensitive primers |
|
S2 EcoR1 |
5´-ACCGACGTCGACTATCCATGAAGAATTCG-3´ |
|
|
S3 EcoR1 |
5´-ACCGACGTCGACTATCCATGAAGAATTCA-3´ |
|
|
S4 EcoR1 |
5´-ACCGACGTCGACTATCCATGAAGAATTCT-3´ |
|
|
S1 MboI |
5´-CACTATCCAGACTCTCACCGCAGATCC-3´ |
|
|
S2 MboI |
5´-CACTATCCAGACTCTCACCGCAGATCG-3´ |
|
|
S3 MboI |
5´-CACTATCCAGACTCTCACCGCAGATCA-3´ |
|
|
S4 MboI |
5´-CACTATCCAGACTCTCACCGCAGATCT-3´ |
|
|
GAPDH F |
5´-GCATGTGCTGACAAAGGAGA-3´ |
Housekeeping gene primers |
|
GAPDH R |
5´-GGTCGTACTCGGGATGATGT-3´ |
|
|
Ubiquitin F |
5´-ATCACGCGCATCTCTCCTAC-3´ |
Target gene primers |
|
Ubiquitin R |
5´-GTGGTGGTGTGGAGTCGAG-3´ |
|
|
AAP3 F |
5´-GGTGTCATCTCGAACTGCTTC-3´ |
|
|
AAP3 R |
5´-CACGAGGTAGATGACAGACATGA-3´ |
|
Table 2.Differentially expressed transcription-derived fragments identified via cDNA-AFLP
Table 2.
|
Code No. |
Accession No. |
Annotation |
Length (bp) |
E-value |
|
1 |
XM-001685021.1 |
L. major amino acid permease (AAP3) gene |
220 |
1e-69 |
|
2 |
XM-003721589.1 |
L. major hypothetical protein gene |
215 |
2e-34 |
|
3 |
Z14232.1 |
L. major ubiquitin gene |
147 |
5e-38 |