Abstract
Diphyllobothrium nihonkaiense has been reported in Korea as Diphyllobothrium latum because of their close morphologic resemblance. We have identified a human case of D. nihonkaiense infection using the mitochondrial cytochrome c oxidase subunit I (cox1) gene sequence analysis. On 18 February 2012, a patient who had consumed raw fish a month earlier visited our outpatient clinic with a long tapeworm parasite excreted in the feces. The body of the segmented worm was 2 m long and divided into the scolex (head) and proglottids. It was morphologically close to D. nihonkaiense and D. latum. The cox1 gene analysis showed 99.4% (340/342 bp) homology with D. nihonkaiense but only 91.8% (314/342 bp) homology with D. latum. The present study suggested that the Diphyllobothrium spp. infection in Korea should be analyzed with specific DNA sequence for an accurate species identification.
-
Key words: Diphyllobothrium nihonkaiense, Diphyllobothrium latum, DNA, sequence, cox1, diagnosis
INTRODUCTION
Most cases of diphyllobothriasis in Korea were reported to have been caused by
Diphyllobothrium latum based on morphologic features of the specimens [
1], which are closely similar to
Diphyllobothrium nihonkaiense. However, genetic analysis can differentiate the 2 species [
2,
3]. In the Republic of Korea (=Korea), through a DNA analysis, 62 diphyllobothriasis cases previously reported as
D. latum infection were verified to have been caused by
D. nihonkaiense [
3]. In addition, the 4
D. latum cases reported in 2012 [
4], which were been verified based on DNA sequencing analysis, may also have been
D. nihonkaiense cases.
It is difficult for clinicians to diagnose correctly the worms with naked eyes. Although the chemotherapy regimen to treat a variety of Diphyllobothrium spp. is the same, it is necessary to identify the species correctly to know the epidemiological characteristics and to prevent further infections by providing information on intermediate hosts. In the present study, a molecular diagnosis of Diphyllobothrium spp. was done using the mitochondrial cytochrome c oxidase subunit I (cox1) gene sequence of the tapeworm discharged from a Korean female patient.
CASE RECORD
A female patient from Chuncheon, Gangwon-do, Korea who ate halibut, Matsubara's rock fish, trout, squid, sea squirts from Sokcho, Gangwon-do, and cherry salmon from Hwacheon, Gangwon-do, 12 months before visiting our clinic on 18 February 2012. She experienced abdominal discomfort for a month before presenting to our clinic [
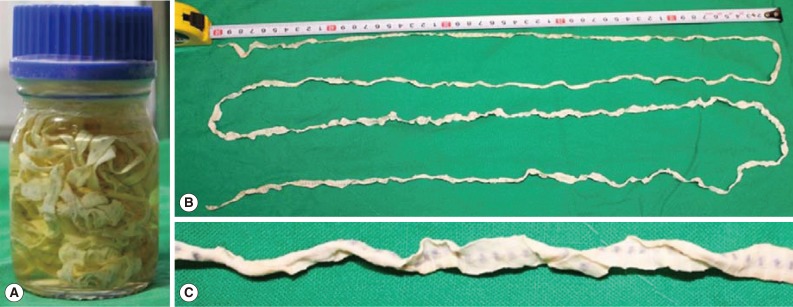
5]. Two days prior to the admission, she experienced abdominal discomfort again, and a part of the tapeworm was observed during the bowel movement. Her husband drew out of the 2 m long worm successfully, which was delivered to our clinic (
Fig. 1).
The results of blood tests were within normal range, and no parasite eggs were observed in the feces during the outpatient follow-up. An abdominal examination did not reveal any atypical presentations. Also no atypical presentation was observed on gastroduodenal endoscopy and colonoscopy performed 2 days after the first visit. The worm specimen was fixed in 10% formalin and sent to the Department of Parasitology, College of Medicine, Hallym University, Chuncheon, Korea. The proglottids of the worm showed a rosette-shaped uterus. The scolex was spoon-shaped with 2 sucking grooves. Therefore, it was identified as Diphyllobothrium spp. on the basis of the morphological finding. The specimen was dispatched to the Department of Parasitology, College of Medicine, Chungbuk National University to identify the species with molecular analysis. The purified PCR-amplified fragments of the cox1 gene were directly sequenced. The primer walking method was employed to obtain direct sequences from each of the amplified fragments. Cyclic sequencing from both ends of the fragments was performed using a Big-Dye Terminator sequencing kit (Applied Biosystems, Foster City, California, USA) and the reaction products were electrophoresed on an automated DNA sequencer (model 3739KL, Applied Biosystems). The sequences were assembled and aligned using Geneous 6.1.5 (Biomatter, Auckland, New Zealand). The sequence regions were identified using BLAST searches and comparisons with sequences of D. nihonkaiense and D. latum, which had been deposited in the GenBank database. The PCR amplification and direct sequencing for the cox1 target fragment (342 bp in length corresponding to the positions 781-1,122 bp of the cox1 gene) were performed using the total genomic DNA extracted from this specimen. The cox1 sequences (342 bp) of the worm showed 99.4% (340/342 bp) similarity to the reference sequence of the Japanese origin D. nihonkaiense (GenBank No. AB015755) and 91.8% (314/342 bp) similarity with the reference sequence of the Russian origin D. latum (GenBank No. DQ985706).
Praziquantel 10 mg/kg in a single dose was administered, and the patient developed headache after taking the drug but was relived soon. The patient's husband was also given the medication because he had eaten the same sea fish with the patient.
DISCUSSION
D. nihonkaiense had been frequently misinterpreted as
D. latum in Korea. In our case, the
cox1 gene sequencing analysis revealed it as
D. nihonkaiense. The diphyllobothriasis reported after 2009 in Korea should be reconfirmed by DNA analysis, and if the same results as ours are obtained, then the
D. latum cases should be revised as those caused by
D. nihonkaiense [
3]. The current case report is also expected to be a resource to aid in
D. nihonkaiense epidemiology. Therefore, we suggest a possibility that
D. latum might not exist in Korea. Although the known intermediate hosts of
D. nihonkaiense are
Oncorhynchus keta and
O. masou, which thrive in the Pacific Ocean [
6], our patient had no history of consuming these kinds of fish. Thus, our case report could prompt further research to discover other intermediate hosts.
References
- 1. Lee EB, Song JH, Park NS, Kang BK, Lee HS, Han YJ, Kim HJ, Shin EH, Chai JY. A case of Diphyllobothrium latum infection with a brief review of diphyllobothriasis in the Republic of Korea. Korean J Parasitol 2007;45:219-223.
- 2. Wicht B, de Marval F, Peduzzi R. Diphyllobothrium nihonkaiense (Yamane et al., 1986) in Switzerland: first molecular evidence and case reports. Parasitol Int 2007;56:195-199.
- 3. Jeon HK, Kim KH, Huh S, Chai JY, Min DY, Rim HJ, Eom KS. Morphologic and genetic identification of Diphyllobothrium nihonkaiense in Korea. Korean J Parasitol 2009;47:369-375.
- 4. Choi HJ, Lee J, Yang HJ. Four human cases of Diphyllobothrium latum infection. Korean J Parasitol 2012;50:143-146.
- 5. Lee SH, Chai JY, Hong ST, Sohn WM, Huh S, Cheong EH, Kang SB. Seven cases of Diphyllobothrium latum infection. Korean J Parasitol 1989;27:213-216. (in Korean).
- 6. Scholz T, Garcia HH, Kuchta R, Wicht B. Update on the human broad tapeworm (genus Diphyllobothrium), including clinical relevance. Clin Microbiol Rev 2009;22:146-160.
Fig. 1Gross findings (A-C) of a spontaneously discharged strobila of D. nihonkaiense from this case. The whole length of this worm was about 2 m. (A, B) Whole discharged worm. (C) A close-up view of the gravid segment.

Citations
Citations to this article as recorded by

- Dietary footprints of a global parasite: diagnosing Dibothriocephalus nihonkaiensis in non-endemic regions
Wilson G.W. Goh, Jean-Marc Chavatte, Gabriel Z.R. Yan, Yuan Yi Constance Chen, Mark Dhinesh Muthiah, Lionel H.W. Lum
Gut Pathogens.2025;[Epub] CrossRef - Repertory of eukaryotes (eukaryome) in the human gastrointestinal tract: taxonomy and detection methods
I. Hamad, D. Raoult, F. Bittar
Parasite Immunology.2016; 38(1): 12. CrossRef - Diphyllobothrium nihonkaiense: wide egg size variation in 32 molecularly confirmed adult specimens from Korea
Seoyun Choi, Jaeeun Cho, Bong-Kwang Jung, Deok-Gyu Kim, Sarah Jiyoun Jeon, Hyeong-Kyu Jeon, Keeseon S. Eom, Jong-Yil Chai
Parasitology Research.2015; 114(6): 2129. CrossRef - Three Cases of Diphyllobothrium nihonkaiense Infection in Korea
Hong-Ja Kim, Keeseon S. Eom, Min Seo
The Korean Journal of Parasitology.2014; 52(6): 673. CrossRef - Extracorporeal Worm Extraction of Diphyllobothrium nihonkaiense with Amidotrizoic Acid in a Child
Hye Kyung Shin, Joo-Hyung Roh, Jae-Won Oh, Jae-Sook Ryu, Youn-Kyoung Goo, Dong-Il Chung, Yong Joo Kim
The Korean Journal of Parasitology.2014; 52(6): 677. CrossRef - Two Human Cases of Diphyllobothrium nihonkaiense Infection in Korea
Su-Min Song, Hye-Won Yang, Min Kyu Jung, Jun Heo, Chang Min Cho, Youn-Kyoung Goo, Yeonchul Hong, Dong-Il Chung
The Korean Journal of Parasitology.2014; 52(2): 197. CrossRef - Parasitic Infections Based on 320 Clinical Samples Submitted to Hanyang University, Korea (2004-2011)
Sung-Chul Choi, Soo-Young Lee, Hyun-Ouk Song, Jae-Sook Ryu, Myoung-Hee Ahn
The Korean Journal of Parasitology.2014; 52(2): 215. CrossRef