Abstract
Mitochondrial genomes have been extensively studied for phylogenetic purposes and to investigate intra- and interspecific genetic variations. In recent years, numerous groups have undertaken sequencing of platyhelminth mitochondrial genomes. Haplorchis taichui (family Heterophyidae) is a trematode that infects humans and animals mainly in Asia, including the Mekong River basin. We sequenced and determined the organization of the complete mitochondrial genome of H. taichui. The mitochondrial genome is 15,130 bp long, containing 12 protein-coding genes, 2 ribosomal RNAs (rRNAs, a small and a large subunit), and 22 transfer RNAs (tRNAs). Like other trematodes, it does not encode the atp8 gene. All genes are transcribed from the same strand. The ATG initiation codon is used for 9 protein-coding genes, and GTG for the remaining 3 (nad1, nad4, and nad5). The mitochondrial genome of H. taichui has a single long non-coding region between trnE and trnG. H. taichui has evolved as being more closely related to Opisthorchiidae than other trematode groups with maximal support in the phylogenetic analysis. Our results could provide a resource for the comparative mitochondrial genome analysis of trematodes, and may yield genetic markers for molecular epidemiological investigations into intestinal flukes.
-
Key words: Haplorchis taichui, trematode, mitochondrial genome, molecular phylogeny
INTRODUCTION
The intestinal trematode,
Haplorchis taichui, is a medically important parasite infecting humans and livestock. Haplorchiasis is a major public health threat in Asia and in parts of Africa and the Americas [
1-
3].
H. taichui is the most frequently reported species among the minute intestinal flukes from Southeast Asia, including Thailand, Lao PDR, China, and Vietnam [
3,
4-
7]. Mitochondrial (mt) genomes exhibit a relatively conserved suite of protein-coding sequences, but also relatively rapid rates of evolutionary change [
8,
9]. In recent years, complete mitochondrial DNA (mtDNA) sequences have been extensively used to infer higher level phylogenies [
10,
11] and also for taxonomy and population genetics at lower taxonomic levels [
12-
14]. To date, quite a number of complete mt genomes of metazoan species, including helminths, have been deposited in GenBank and published [
15]. Information from flatworm mitochondrial genomes is strongly biased toward parasitic species of medical importance. For this reason, recent mitochondrial genome scale phylogenetic surveys have emphasized the need to collect data for the major groups of flatworms that have not been sampled [
16,
17]. However, most of them still remain poorly understood at the molecular level, in particular, the complete mt genomes of the species in the family Heterophyidae. Parasitic flatworm mt genomes, ranging in size usually from 13 to 14 kb but far bigger up to 24 kb sometimes, are typically circular and usually encode 36 genes, including 12 protein-coding genes, and without introns and with short intergenic regions [
18]. The Digenea currently contains about 18,000 nominal species parasitizing vertebrates, and sometimes humans as the definitive host [
19]. The purpose of the present study was to sequence the mt genome of
H. taichui for comparison with the organization and sequence of the mt genomes of other trematodes. In addition, we wished to reconstruct the phylogenetic relationships of the family Heterophyidae within the class Trematoda using mtDNA sequences.
MATERIALS AND METHODS
Long PCR amplification and sequencing of the H. taichui mtDNA molecule
Adult
H. taichui worms were obtained from naturally infected Laotian people during the activity of "Korea-Lao PDR Collaborative Project for Control of Foodborne-Trematode Infections (esp. Opisthorchiasis) in Lao PDR (2007-2011)". The specimens were washed in normal physiological saline and identified based on morphological characters (gonotyl bears 12-16 spines in
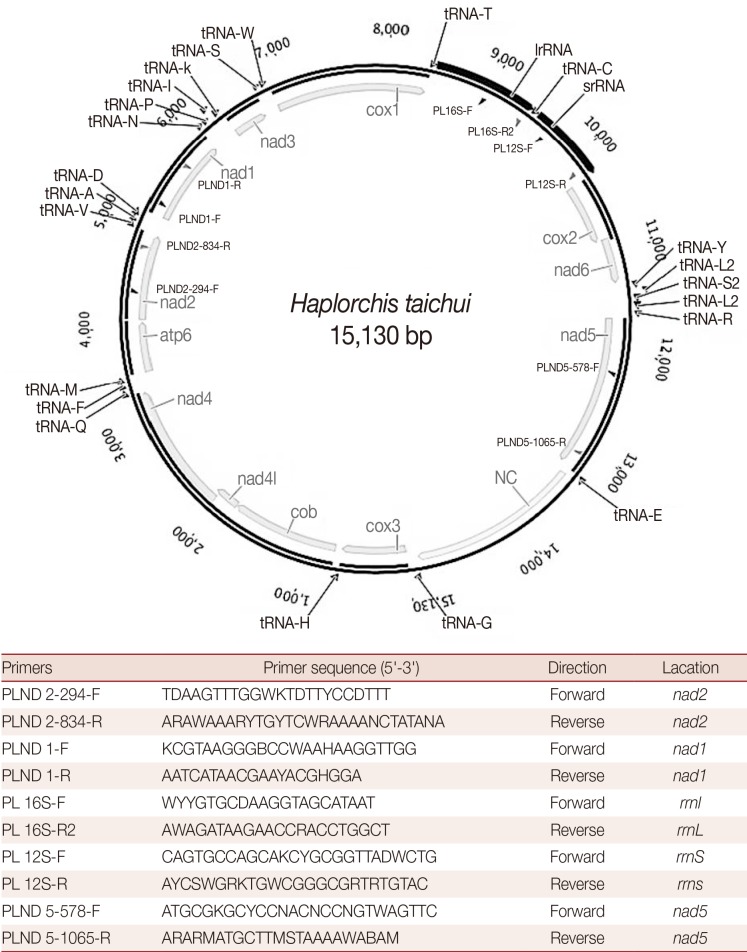
H. taichui). The worms were stored in 70% ethanol prior to DNA extraction. Total genomic DNA was extracted from 200 specimens using a QIAamp tissue kit (Qiagen Inc., Valencia, California, USA), according to the manufacturer's instruction and used as the template DNA. The complete mt genome was PCR-amplified in overlapping fragments. The nucleotide sequences and relative positions of the PCR primers are shown in
Fig. 1. PCR reactions were performed in a 50 µl reaction volume consisting of 10 units of EF-Taq polymerase (Solgent Co., Daejeon, Korea), 2.5 mM dNTP mixture, 2.5 mM MgCl
2, 20
pmole of each primer, and 10 µg of genomic DNA in a thermocycler (Biometra Co., Goettingen, Germany) under the following conditions: 92℃ for 2 min (initial denaturation), then 92℃ for 10 sec (denaturation), 50℃ for 30 sec (annealing), and 68℃ for 2 min to 10 min (extension) for 10 cycles, followed by 92℃ for 2 min, then 92℃ for 10 sec, 48℃ for 30 sec, and 68℃ for 2 min to 10 min for 20 cycles, and a final extension at 68℃ for 8 min.
A negative control (no template) was also included for every PCR reaction to detect contamination. Each amplicon (3 µl) was examined by agarose (1%) gel electrophoresis, stained with ethidium bromide and photographed using a gel documentation system (UVItec, Cambridge, UK), excised under long-wavelength UV light, extracted using a Doc-do purification kit (Elpis Co., Daejeon, Korea), and then used as a template for sequencing reactions. The primer walking method was employed to obtain overlapping sequences for each of the amplified fragments. Cyclic sequencing from both ends of the fragments was performed with a Big-Dye, and the amplified products were subjected to electrophoresis on an ABI 3100 automated DNA sequencer.
Sequence analysis and characterization of the Haplorchis taichui mt genome
Gene annotation, genome organization, translation initiation, translation termination codons, and the boundaries between protein-coding genes of mt genomes were identified based on comparison with mt genomes of other trematodes reported previously [
17,
18]. Sequences were assembled manually and aligned against the complete mt genome sequences of our own
Metagonimus yokogawai sequence (will be published elsewhere) and trematode parasites available in the GenBank database (
http://www.ncbi.nlm.nih.gov WebGenBank) using BLAST searches. Open reading frames and gene boundaries were confirmed by comparing with
M. yokogawai nucleotide sequences. The codon usage profiles of 12 protein-coding genes and their nucleotide composition were calculated using Geneious 6.1.5 (Biomatters Co., Auckland, New Zealand) program [
20]. Putative secondary structures of 22 tRNA genes were identified manually by recognizing potential secondary structures and anticodon sequences.
To assess the phylogenetic position of
H. taichui and the utility of mt genomes in resolving the interrelationships of trematode orders, complete mitochondrial genome sequences of 15 flatworms were analyzed. The mtDNA sequences were as follows:
H. taichui (KF214770),
Trichobilharzia regenti (NC_009680),
Schistosoma spindale (DQ157223),
S. haematobium (DQ157222),
S. mansoni (NC_002545),
S. japonicum (NC_002544),
S. mekongi (NC_002529),
Fasciola hepatica (NC_002546),
Paragonimus westermani (NC_002354),
Opisthorchis felineus (EU921260),
O. viverrini (JF739555),
Clonorchis sinensis (FJ381664/JF729303/JF729304), and 1 Monogenea species (
Gyrodactylus thymalli, NC_009682) as the outgroup. Each gene was translated into an amino acid sequence using the trematode mt genetic code by translation table 21 in Geneious 6.1.5, and was aligned based on its amino acid sequence using default settings. A conserved block of concatenated alignment was selected using the Gblocks program [
21], (
http://molevol.cmima.csic.es/castresana/Gblocks_server) for 12 protein-coding genes examined. Phylogenetic relationships among trematodes were inferred using different tree construction algorithms, i.e., maximum parsimony (MP), neighbor-joining (NJ), maximum likelihood (ML), Bayesian phylogeny (BP) [
22], PAUP
* [
23], PhyML 3.0 [
24], and MrBayes [
25]. Bootstrap analysis was done using 1,000 random replications. Models of amino acid substitution were determined for each data partition independently using ModelTest supported in Geneious 6.1.5. The MP analysis was performed using the exhaustive search option. ML analysis was performed using the substitution model Le Gascuel (LG). In BP analysis, the following settings were applied: the number of cycles=1,100,000; sampling frequency=200; heated chains=4; burn-in=100,000.
RESULTS
General features of the mt genome of H. taichui
The complete mt genome of
H. taichui Lao PDR isolate (GenBank accession no. KF214770) is 15,130 bp in length (
Fig. 1). It encodes 36 genes; 12 protein-coding genes (
cox1-3,
nad1-6,
nad4L,
atp6,
cob, and lacking
atp8), 22 transfer RNA genes, and 2 ribosomal RNA genes.The relative positions and lengths of each gene are given in
Table 1. An AT-rich region (1,710 bp) is located between
tRNA-Glu and
tRNA-Gly. All genes are transcribed in the same direction. The 2 adjacent genes,
nad4L and
nad4, overlap each other by 40 nt in different reading frames.
The mt genome of
H. taichui encodes 12 protein-coding genes, identical with the situation in other trematodes. The start and termination codons of these were identified by sequence comparison with homologs in other trematodes. The ATG codon was used in 9 protein-coding genes, and the GTG codon in 3 genes (
nad1,
nad4, and
nad5). The TAG stop codon was used in 9 genes (
cox3,
cob,
nad4L,
atp6,
nad2,
nad1,
nad3,
cox1, and
nad6) and the TAA termination codon in the remaining 3 genes (
nad4,
cox2, and
nad5). Pairwise comparisons were made among the amino acid sequences inferred from individual protein-coding genes in the
H. taichui genome with those representing 12 other trematodes (
Table 2). The amino acid sequence similarities in individual inferred proteins ranged from 76.8% (
cox1) to 82.4% (
cob) between
H. taichui and
O. felineus; and from 78.0% (
cox1) to 80.5% (
cob) between
H. taichui and
C. sinensis. The amino acid sequence similarities between
H. taichui and
S. japonicum ranged from 24.2% (
cox3) to 67.9% (
cox1); and from 47.1% (
nad2) to 75.6% (
cob) with
F. hepatica (
Table 2). The 12 protein-coding genes were 10,176 bp in length and composed of 43% T, 17.1% A, 28% G, and, 11.9% C, accounting for 67.3% of the full length of the genome (
Table 3). All 64 codons were used (
Table 4). However, some codons, such as CGC, CGA for arginine, ACA for threonine, or GCA for alanine, were very uncommon, reflecting the nucleotide composition. Several amino acids, histidine (1.6%), lysine (1.3%), and glutamine (0.7%), were rarely used. Five of the most frequently used amino acids were leucine (16.7%), valine (11.9%), serine (10.4%), phenylalanine (9.9%), and glycine (9.0%). These collectively constituted 57.9% of the total number of amino acids. Amino acids encoded by T-rich codons (≥ 2 Ts in a triplet) were the most abundant and accounted for 42.7% of the total amino acid composition, whereas C-rich codons (≥ 2 Cs in a triplet) were the least used (they accounted for merely 5.0% of the total amino acid composition). As shown in
Table 4, unequal usage of synonymous codons avoiding C at the third codon position was prominent in most cases; for instance, the relative frequency of using TTT for Phe was 9.2%, but the frequency of using TTC was only 1.4%.
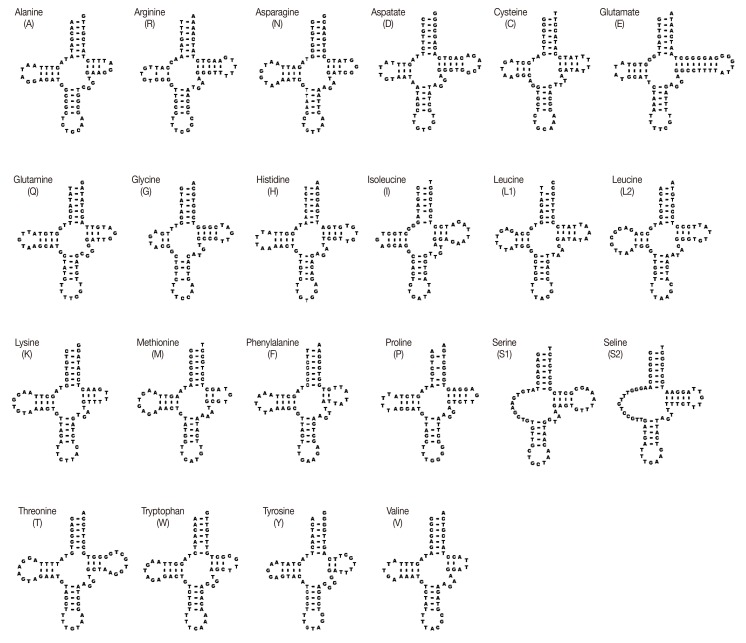
The sizes of 22 tRNA genes identified in mt genomes of
H. taichui ranged from 60 to 73 nucleotides (nt) in length. Of the 22 tRNA genes, 20 could be folded into the conventional cloverleaf structure, including a 7 bp amino-acyl stem, a 2-4 bp DHU arm with a 3-10 nt loop, a 5 bp anticodon stem with a loop of 7 nt, and a 2-8 bp of the TψC arm with a 3-10 nt loop. The 2 tRNAs specifying serine were exceptions (
Fig. 2). The
rrnL was located between
tRNA-Thr and
tRNA-Cys, and
rrnS was located between
tRNA-Cys and
cox2. The
rrnS and
rrnL of
H. taichui were 747 nt and 979 nt in length. Each ribosomal gene was assumed to directly abut neighboring genes. The A+T contents of the
rrnL was 58.5%, respectively, and the A+T content of the
rrnS was 55.9%. Just 1 long non-coding region (LNR) was identified, located between the
tRNA-Gly and
tRNA-Glu, and lacked any tandem repeats. The size was 1,710 bp, comprising 11.3% of the genome, and the A+T content was 58.3%.
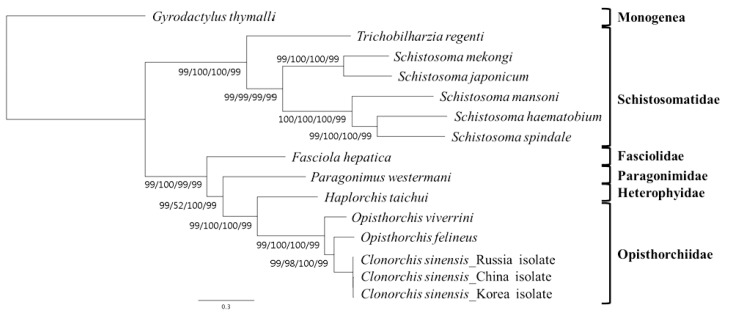
A concatenated alignment set of 3,380 homologous amino acid positions from conserved blocks was used. Phylogenetic relationships among the 15 flatworms using different analytical approaches (MP, ML, NJ, and BP methods) were the same in their topology (
Fig. 3). Phylogenetic relationships among species were well resolved with very high nodal support throughout. In this tree, Schistosomatidae and (Fasciolidae+Paragonimidae+Opisthorchiidae+Heterophyidae) formed monophyletic groups.
H. taichui was resolved as sister to Opisthorchiidae with a very high support in the phylogenetic analysis.
DISCUSSION
The mt genome arrangement of
H. taichui was the same as that of
Fasciola hepatica (NC_002546),
Paragonimus westermani (NC_002354) and
Opisthorchis spp., but distinct from the arrangement seen in some
Schistosoma spp. [
9,
18]. All genes were transcribed in the same direction, as in other flatworms for which data are available [
26].
H. taichui lacks
atp8, a gene that is not seen in any flatworm species [
18]. The majority of metazoan mtDNA sequences contain 2 non-coding regions of significant size difference, a long non-coding region (LNR) and a short non-coding region (SNR). In the case of
H. taichui, a single long non-coding region was present. The nucleotide composition of the entire mtDNA of
H. taichui was biased toward T and G, with T being the most common nucleotide and C the least favored, in accordance with mt genomes of other trematodes except for
P. westermani (
Table 3). Two genes,
nad4L and
nad4, overlapped by 40 nucleotides (
Table 1), similar to the situation in other digeneans. The tRNA genes generally resemble those of other digeneans. A standard cloverleaf structure can be inferred for most tRNAs. Exceptions are tRNA-S in which the paired dihydrouridine (DHU) arm is missing in all parasitic flatworm species [
18], although secondary structures including this arm are feasible for some species [
15] including
H. taichui. The
rrnS was 747 nt in length, shorter than that of the homologs from other trematodes except for
S. mansoni (744 nt) and
P. westermani (744 nt, not registered on GenBank). The rrnS of
S. mekongi was noted as being 709 nt (GenBank accession no. NC_002529), but it could be 39 nt longer if it directly abuts
cox2 in that species. The
rrnL of
H. taichui, at 979 nt, was the shortest among trematodes recorded yet. Morphological data have traditionally been used for taxonomic studies on flatworms. Such data are now being supplemented by data from ultra-structural and biochemical studies [
27,
28] and, increasingly, from molecular sequences. Published phylogenies using nuclear ribosomal genes [
19,
29] indicate that the Heterophyidae is paraphyletic with respect to the Opisthorchiidae. Our mitochondrial sequences have yielded a tree consistent with this finding, with
H. taichui seen as a sister to
Clonorchis+
Opisthorchis (family Opisthorchiidae). Sequences from additional heterophyid and opisthorchiid mt genomes will be required to confirm the findings from nuclear genes.
In conclusion, the present study reported the complete mtDNA sequence and genome organization of H. taichui for the first time. Its constituent genes were compared with homologs from other trematodes. Our phylogenetic analysis of concatenated protein-coding genes supports a sister group relationship between families Opisthorchiidae and Heterophyidae. These data will provide tools for the molecular diagnosis of haplorchiasis and for studies on the biology of the species.
National Research FoundationNRF-2010-0024745
ACKNOWLEDGMENTS
The Parasite Resource Bank of Korea National Research Resource Center, Republic of Korea, provided the materials for morphological identification. The National Research Foundation of Korea (NRF-2010-0024745) financially supported this work.
References
- 1. Chai JY, Murrell KD, Lymbery AJ. Fish-borne parasitic zoonoses: status and issues. Int J Parasitol 2005;35:1233-1254.
- 2. Trung Dung D, De NV, Waikagul J, Dalsgaard A, Chai JY, Sohn WM, Murrell KD. Fishborne zoonotic intestinal trematodes, Vietnam. Emerg Infect Dis 2007;13:1828-1833.
- 3. Chai JY, Shin EH, Lee SH, Rim HJ. Foodborne intestinal flukes in Southeast Asia. Korean J Parasitol 2009;47(suppl):S69-S102.
- 4. Thaenkham U, Visetssuk K, Dung DT, Waikagul J. Discrimination of Opisthorchis viverrini from Haplorchis taichui using COI sequence marker. Acta Trop 2007;103:26-32.
- 5. Jeon HK, Lee DM, Park HS, Min DY, Rim HJ, Zhang H, Yang Y, Li X, Eom KS. Human infections with liver and minute intestinal flukes in Guangxi, China: analysis by DNA sequencing, ultrasonography, and immunoaffinity chromatography. Korean J Parasitol 2012;50:391-394.
- 6. Chai JY, De NV, Sohn WM. Foodborne trematode metacercariae in fish from northern Vietnam and their adults recovered from experimental hamsters. Korean J Parasitol 2012;50:317-325.
- 7. Wongsawad C, Wongsawad P, Chuboon S, Anuntalabhochai S. Coprodiagnosis of Haplorchis taichui infection using sedimentation and PCR-based methods. Southeast Asian J Trop Med Public Health 2009;40:924-928.
- 8. Jex AR, Waeschenbach A, Littlewood DTJ, Hu M, Gasser RB. The mitochondrial genome of Toxocara canis. PLos Negl Trop Dis 2008;2:e273.
- 9. Lawton SP, Hirai H, Elronside JE, Johnston DA, Rollinson D. Genomes and geography: genomic insights into the evolution and phylogeography of the genus Schistosoma. Parasit Vectors 2011;4:131.
- 10. Cavalier-Smith T. Protist phylogeny and the high-level classification of Protozoa. Eur J Protistol 2003;39:338-348.
- 11. Gissi C, Iannelli F, Pesole G. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity (Edinb) 2008;101:301-320.
- 12. Jeon HK, Kim KH, Eom KS. Complete sequence of the mitochondrial genome of Taenia saginata: comparison with T. solium and T. asiatica. Parasitol Int 2007;56:243-246.
- 13. Park JK, Kim KH, Kang S, Jeon HK, Kim JH, Littlewood DTJ, Eom KS. Characterization of the mitochondrial genome of Diphyllobothrium latum (Cestoda: Pseudophyllidea)-implications for the phylogeny of eucestodes. Parasitology 2007;134:749-759.
- 14. Liu GH, Wu CY, Song HQ, Wei SJ, Xu MJ, Lin RQ, Zhao GH, Huang SY, Zhu XZ. Comparative analyses of the complete mitochondrial genomes of Ascaris lumbricoides and Ascaris suum from humans and pigs. Gene 2012;492:110-116.
- 15. Shekhovtsov SV, Katokhin AV, Kolchanov NA, Mordvinov VA. The complete mitochondrial genomes of the liver flukes Opisthorchis felineus and Clonorchis sinensis (Trematoda). Parasitol Int 2010;59:100-103.
- 16. Kim KH, Jeon HK, Kang S, Sultana T, Kim GJ, Eom KS, Park JK. Charaterization of the complete mitochondrial genome of Diphyllobothrium nihonkaiense (Diphyllobothriidae: Cestoda), and Development of molecular markers for differentiating fish tapeworms. Mol Cells 2007;23:379-390.
- 17. Cai XQ, Liu GH, Song HQ, Wu CY, Zou FC, Yan HK, Yuan ZG, Lin RQ, Zhu XQ. Sequences and gene organization of the mithochondrial genomes of the liver flukes Opisthorchis viverrini and Clonorchis sinensis (Trematoda). Parasitol Res 2012;110:235-243.
- 18. Le TH, Blair D, McManus DP. Mitochondrial genomes of parasitic flatworms. Trends Parasitol 2002;18:206-213.
- 19. Olson PD, Cribb TH, Tkach VV, Bray RA, Littlewood DTJ. Phylogeny and classification of Digenea (Platyhelminthes: Trematoda). Int J Parasitol 2003;33:733-755.
- 20. Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meinthes P, Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012;28:1647-1649.
- 21. Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 2007;56:564-577.
- 22. Masters BC, Fan V, Ross HA. Species delimitation-a geneious plugin for the species boundaries. Mol Ecol Resour 2011;11:154-157.
- 23. Swofford DL. Paup*: phylogenetic analysis using parsimony (* and other methods). Version 4. Sunderland, Massachusetts, USA. Sinauer Associates; 2003.
- 24. Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 2003;52:696-704.
- 25. Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 2001;17:754-755.
- 26. Johnston DA. Genomes and genomics of parasitic flatworms. In Maule AG, Marks NJ eds, Parasitic Flatworms: Molecular Biology, Biochemistry, Immunology and Physiology. Wallingford, Oxford, UK. CAB International; 2006, pp 37-80.
- 27. Justine JL. Phylogeny of parasitic Platyhelminthes: a critical study of synapomorphies proposed on the basis of the ultrastructure of spermiogenesis and spermatozoa. Can J Zool 1991;69:1421-1440.
- 28. Jeon HK, Eom KS. Immunoblot patterns of Taenia asiatica taeniasis. Korean J Parasitol 2009;47:73-77.
- 29. Thaenkham U, Blair D, Nawa Y, Waikagul J. Families Opisthorchiidae and Heterophyidae: are they distinct? Parasitol Int 2012;61:90-93.
Fig. 1A map of the complete mitochondrial genome of Haplorchis taichui. The primer sequences used for amplification of respective mitochondrial genes are indicated in the map.

Fig. 2DNA sequences for 22 tRNA genes of H. taichui mtDNA folded into inferred secondary structures.
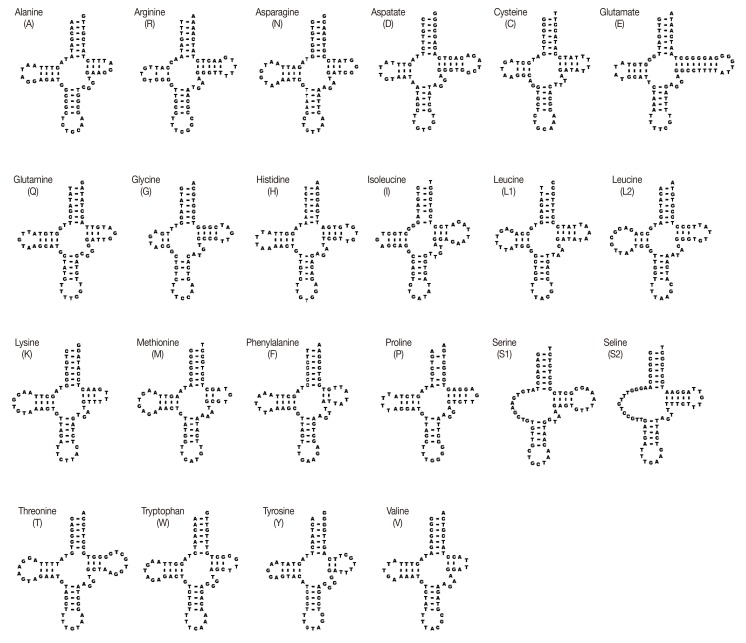
Fig. 3Phylogenetic relationships among trematode parasites based on inferred amino acid sequences of 12 mitochondrial protein-coding gene loci for 11 species 14 individual using the outgroup, G. thymalli (GenBank accession no. NC_009682) by MP/ML/NJ/BP, respectively (branch length for ML).
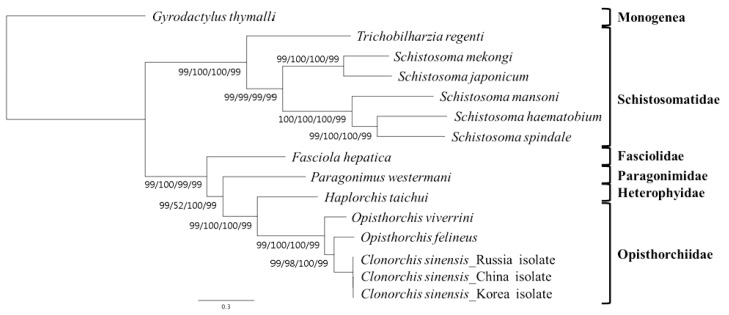
Table 1.Position and characteristics of protein-coding and non-coding sequences in the mt genome of Haplorchis taichui
Table 1.
|
Gene/Region |
No. of
|
Codons
|
Positions
|
|
Nucleotides |
Amino acids |
Initiation |
Termination |
(5´-3´) |
|
cox3
|
657 |
218 |
ATG |
TAG |
1-657 |
|
tRNA-H
|
66 |
|
|
|
660-725 |
|
cob
|
1,110 |
369 |
ATG |
TAG |
732-1841 |
|
nad4L
|
264 |
87 |
ATG |
TAG |
1843-2106 |
|
nad4
|
1,281 |
426 |
GTG |
TAA |
2067-3347 |
|
tRNA-Q
|
66 |
|
|
|
3357-3422 |
|
tRNA-F
|
62 |
|
|
|
3427-3488 |
|
tRNA-M
|
64 |
|
|
|
3489-3552 |
|
atp6
|
516 |
171 |
ATG |
TAG |
3553-4068 |
|
nad2
|
870 |
289 |
ATG |
TAA |
4094-4963 |
|
tRNA-V
|
60 |
|
|
|
4968-5027 |
|
tRNA-A
|
63 |
|
|
|
5029-5091 |
|
tRNA-D
|
67 |
|
|
|
5094-5160 |
|
nad1
|
906 |
301 |
GTG |
TAG |
5161-6066 |
|
tRNA-N
|
66 |
|
|
|
6069-6134 |
|
tRNA-P
|
64 |
|
|
|
6139-6202 |
|
tRNA-I
|
65 |
|
|
|
6199-6263 |
|
tRNA-K
|
65 |
|
|
|
6267-6331 |
|
nad3
|
360 |
119 |
ATG |
TAG |
6332-6691 |
|
tRNA-S
|
62 |
|
|
|
6705-6766 |
|
tRNA-W
|
64 |
|
|
|
6774-6837 |
|
cox1
|
1,542 |
513 |
ATG |
TAG |
6842-8383 |
|
tRNA-T
|
61 |
|
|
|
8385-8445 |
|
rrnL
|
979 |
|
|
|
8446-9424 |
|
tRNA-C
|
64 |
|
|
|
9426-9489 |
|
rrnS
|
747 |
|
|
|
9490-10236 |
|
cox2
|
624 |
207 |
ATG |
TAG |
10237-10860 |
|
nad6
|
459 |
152 |
ATG |
TAG |
10847-11305 |
|
tRNA-Y
|
67 |
|
|
|
11306-11372 |
|
tRNA-L1
|
66 |
|
|
|
11370-11435 |
|
tRNA-S2
|
64 |
|
|
|
11434-11497 |
|
tRNA-L2
|
70 |
|
|
|
11504-11573 |
|
tRNA-R
|
64 |
|
|
|
11585-11648 |
|
nad5
|
1,587 |
528 |
GTG |
TAA |
11650-13236 |
|
tRNA-E
|
73 |
|
|
|
13248-13320 |
|
NR
|
1,710 |
|
|
|
13323-15032 |
|
tRNA-G
|
59 |
|
|
|
15035-15093 |
Table 2.Properties of trematode mtDNA protein-coding genes and similarity comparison between Haplorchis taichui and other trematodes
Table 2.
|
Gene |
Species
|
|
H.t |
S.s |
S.h |
S.m |
S.j |
S.ml |
S.mk |
F.h |
P.w |
O.f |
O.v |
C.s |
|
No. of amino acids |
|
|
|
|
|
|
|
|
|
|
|
|
|
cox3
|
218 |
221 |
221 |
218 |
215 |
217 |
217 |
214 |
215 |
214 |
214 |
213 |
|
cob
|
369 |
364 |
367 |
365 |
372 |
373 |
373 |
371 |
373 |
371 |
369 |
370 |
|
nad4L
|
87 |
84 |
86 |
87 |
88 |
88 |
88 |
91 |
86 |
87 |
87 |
87 |
|
nad4
|
426 |
420 |
421 |
420 |
425 |
424 |
424 |
424 |
402 |
425 |
425 |
425 |
|
atp6
|
171 |
173 |
174 |
174 |
173 |
174 |
174 |
173 |
171 |
171 |
171 |
171 |
|
nad2
|
289 |
279 |
279 |
280 |
285 |
284 |
284 |
289 |
289 |
289 |
289 |
290 |
|
nad1
|
301 |
291 |
298 |
297 |
297 |
298 |
296 |
301 |
297 |
300 |
300 |
300 |
|
nad3
|
119 |
122 |
122 |
121 |
120 |
121 |
121 |
119 |
119 |
118 |
118 |
118 |
|
cox1
|
513 |
587 |
601 |
511 |
509 |
> 433 |
511 |
511 |
498 |
520 |
516 |
519 |
|
cox2
|
207 |
200 |
198 |
198 |
203 |
u |
233 |
201 |
200 |
212 |
214 |
211 |
|
nad6
|
152 |
155 |
157 |
150 |
153 |
u |
154 |
151 |
151 |
153 |
153 |
153 |
|
nad5
|
528 |
528 |
527 |
528 |
529 |
> 333 |
531 |
523 |
528 |
534 |
534 |
534 |
|
Amino acid similarity (%) |
|
|
|
|
|
|
|
|
|
|
|
|
|
cox3
|
100 |
28.7 |
28.2 |
27.3 |
24.2 |
30.0 |
28.2 |
49.5 |
54.0 |
49.5 |
50.0 |
49.3 |
|
cob
|
100 |
48.5 |
51.1 |
52.9 |
54.9 |
53.0 |
53.8 |
75.6 |
75.9 |
82.4 |
77.8 |
80.5 |
|
nad4L
|
100 |
32.9 |
322.6 |
32.6 |
32.2 |
31.0 |
33.3 |
64.4 |
67.1 |
72.4 |
73.6 |
71.3 |
|
nad4
|
100 |
34.0 |
30.9 |
32.8 |
29.4 |
30.4 |
31.3 |
50.7 |
52.7 |
59.6 |
59.1 |
58.9 |
|
atp6
|
100 |
38.5 |
34.5 |
33.9 |
39.9 |
41.4 |
38.7 |
62.8 |
58.5 |
64.9 |
66.1 |
67.8 |
|
nad2
|
100 |
32.3 |
30.6 |
33.3 |
32.3 |
33.3 |
34.4 |
47.1 |
46.4 |
51.9 |
53.3 |
52.2 |
|
nad1
|
100 |
41.1 |
40.2 |
43.1 |
46.8 |
42.1 |
43.1 |
69.9 |
69.6 |
72.6 |
70.9 |
70.2 |
|
nad3
|
100 |
32.8 |
37.0 |
37.3 |
37.0 |
41.7 |
37.0 |
63.0 |
60.5 |
67.2 |
62.2 |
63.9 |
|
cox1
|
100 |
68.0 |
68.0 |
68.2 |
67.9 |
nc |
66.5 |
74.7 |
78.5 |
76.8 |
78.9 |
78.0 |
|
cox2
|
100 |
44.6 |
42.6 |
45.8 |
45.4 |
nc |
41.6 |
48.8 |
48.5 |
59.5 |
58.5 |
61.0 |
|
nad6
|
100 |
34.0 |
29. |
34.2 |
35.7 |
nc |
36.4 |
51.0 |
54.0 |
57.2 |
60.5 |
57.2 |
|
nad5
|
100 |
34.0 |
32.8 |
31.2 |
32.5 |
nc |
32.4 |
52.0 |
50.3 |
46.5 |
44.6 |
46.0 |
|
Inferred initiation/termination codona
|
|
|
|
|
|
|
|
|
|
|
|
|
|
cox3
|
A/G |
A/A |
A/G |
G/G |
A/G |
A/A |
A/G |
A/G |
A/G |
A/G |
A/G |
A/G |
|
cob
|
A/G |
A/G |
A/A |
G/G |
A/G |
A/A |
A/A |
A/G |
A/G |
A/G |
A/G |
A/G |
|
nad4L
|
A/G |
A/A |
A/A |
A/A |
A/A |
A/A |
A/A |
G/G |
G/G |
A/G |
A/G |
A/G |
|
nad4
|
G/A |
A/A |
A/G |
A/A |
A/G |
A/A |
A/A |
G/A |
A/G |
A/G |
A/G |
G/G |
|
atp6
|
A/G |
A/A |
A/G |
A/G |
A/A |
A/G |
A/A |
A/G |
A/G |
A/G |
A/G |
A/G |
|
nad2
|
A/A |
A/A |
A/A |
G/A |
A/G |
A/G |
A/A |
A/G |
A/A |
A/G |
A/G |
G/G |
|
nad1
|
G/G |
A/A |
A/G |
G/G |
A/G |
A/A |
A/A |
G/G |
A/G |
G/G |
G/G |
G/G |
|
nad3
|
A/G |
A/A |
A/A |
A/G |
A/G |
A/G |
A/G |
A/G |
A/G |
G/G |
G/G |
G/G |
|
cox1
|
A/G |
A/A |
A/G |
A/G |
G/G |
A/nc |
A/A |
A/G |
A/G |
G/G |
G/G |
G/A |
|
cox2
|
A/G |
A/A |
A/G |
A/A |
A/A |
nc/nc |
A/A |
A/G |
A/G |
A/G |
A/G |
A/G |
|
nad6
|
A/G |
A/G |
A/A |
A/A |
A/G |
nc/nc |
G/A |
A/G |
A/G |
A/G |
A/G |
G/A |
|
nad5
|
G/A |
A/G |
A/G |
A/G |
A/G |
nc/G |
G/A |
G/G |
G/G |
A/G |
A/A |
G/A |
Table 3.Nucleotide content of protein-coding genes from complete or almost complete mitochondrial genomes of flatworms
Table 3.
|
Species |
GenBank accession no. |
Base composition (%)
|
Total bp usage |
Total no. of codons |
|
T |
C |
A |
G |
A+T |
|
Schistosoma spindale
|
DQ157223 |
45.2 |
7.0 |
28.1 |
19.7 |
73.3 |
10,308 |
3,424 |
|
Schistosoma haematobium
|
DQ157222 |
44.9 |
7.6 |
27.8 |
19.5 |
72.7 |
10,389 |
3,451 |
|
Schistosoma mansoni
|
NC_002545 |
45.6 |
8.2 |
23.3 |
23.0 |
68.9 |
10,083 |
3,349 |
|
Schistosoma japonicum
|
NC_002544 |
48.3 |
8.0 |
23.0 |
20.7 |
71.3 |
10,143 |
3,369 |
|
Schistosoma malayensisa
|
AAG60031 |
48.8 |
6.6 |
23.6 |
20.9 |
72.4 |
8,271 |
2,745 |
|
Schistosoma mekongi
|
NC_002529 |
48.4 |
6.7 |
24.3 |
20.6 |
72.7 |
10,254 |
3,406 |
|
Fasciola hepatica
|
NC_002546 |
49.4 |
9.6 |
14.2 |
26.8 |
63.6 |
10,140 |
3,368 |
|
Paragonimus westermani
|
NC_002354 |
38.3 |
17.9 |
13.2 |
30.6 |
51.5 |
10,023 |
3,329 |
|
Opisthorchis felineus
|
EU921260 |
45.3 |
12.1 |
15.3 |
27.2 |
60.6 |
10,217 |
3,394 |
|
Opisthorchis viverrini
|
JF739555 |
44.9 |
12.3 |
15.5 |
27.3 |
60.4 |
10,206 |
3,390 |
|
Clonorchis sinensis
|
FJ381664 |
45.1 |
11.9 |
15.7 |
27.3 |
60.8 |
10,209 |
3,391 |
|
Haplorchis taichui
|
KF214770 |
43.0 |
11.9 |
17.1 |
28.0 |
60.1 |
10,176 |
3,380 |
Table 4.Nucleotide codon usage for 12 protein-encoding genes of the mitochondrial genome of Haplorchis taichui
Table 4.
|
AA |
Codon |
No. |
% |
AA |
Codon |
No. |
% |
AA |
Codon |
No. |
% |
AA |
Codon |
No. |
% |
AA |
Ab |
No. |
% |
|
Phe |
UUU(F) |
308 |
9.19 |
Ser |
UCU(S) |
96 |
2.87 |
Tyr |
UAU(Y) |
136 |
4.06 |
Cys |
UGU(C) |
98 |
3.93 |
Ala |
A |
146 |
4.3 |
|
Phe |
UUC(F) |
48 |
1.43 |
Ser |
UCC(S) |
20 |
0.60 |
Tyr |
UAC(Y) |
35 |
1.04 |
Cys |
UGC(C) |
22 |
0.66 |
Cys |
C |
101 |
3.0 |
|
Leu |
UUA(L) |
145 |
4.33 |
Ser |
UCA(S) |
22 |
0.66 |
*** |
UAA(*) |
3 |
0.09 |
Trp |
UGA(W) |
49 |
1.46 |
Asp |
D |
71 |
2.1 |
|
Leu |
UUG(L) |
204 |
6.09 |
Ser |
UCG(S) |
34 |
1.01 |
*** |
UAG(*) |
9 |
0.27 |
Trp |
UGG(W) |
76 |
2.27 |
Glu |
E |
78 |
2.3 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Phe |
F |
336 |
9.9 |
|
Leu |
CUU(L) |
93 |
2.78 |
Pro |
CCU(P) |
35 |
1.04 |
His |
CAU(H) |
36 |
1.07 |
Arg |
CGU(R) |
34 |
1.01 |
Gly |
G |
303 |
9.0 |
|
Leu |
CUC(L) |
23 |
0.69 |
Pro |
CCC(P) |
16 |
0.48 |
His |
CAC(H) |
8 |
0.24 |
Arg |
CGC(R) |
7 |
0.21 |
His |
H |
54 |
1.6 |
|
Leu |
CUA(L) |
22 |
0.66 |
Pro |
CCA(P) |
13 |
0.39 |
Gln |
CAA(Q) |
8 |
0.24 |
Arg |
CGA(R) |
8 |
0.24 |
Ile |
I |
107 |
3.2 |
|
Leu |
CUG(L) |
52 |
1.55 |
Pro |
CCG(P) |
15 |
0.45 |
Gln |
CAG(Q) |
17 |
0.51 |
Arg |
CGG(R) |
31 |
0.93 |
Lys |
K |
44 |
1.3 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Leu |
L |
565 |
16.7 |
|
Ile |
AUU(I) |
98 |
2.93 |
Thr |
ACU(T) |
47 |
1.40 |
Asn |
AAU(N) |
33 |
0.99 |
Ser |
AGU(S) |
63 |
1.88 |
Met |
M |
186 |
5.5 |
|
Ile |
AUC(I) |
14 |
0.42 |
Thr |
ACC(T) |
11 |
0.33 |
Asn |
AAC(N) |
6 |
0.18 |
Ser |
AGC(S) |
14 |
0.42 |
Asn |
N |
72 |
2.1 |
|
Met |
AUA(M) |
65 |
1.94 |
Thr |
ACA(T) |
9 |
0.27 |
Asn |
AAA(N) |
33 |
0.99 |
Ser |
AGA(S) |
31 |
0.93 |
Pro |
P |
92 |
2.7 |
|
Met |
AUG(M) |
108 |
3.22 |
Thr |
ACG(T) |
16 |
0.48 |
Lys |
AAG(K) |
41 |
1.22 |
Ser |
AGG(S) |
62 |
1.85 |
Gln |
Q |
24 |
0.7 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Arg |
R |
67 |
2.0 |
|
Val |
GUU(V) |
171 |
5.10 |
Ala |
GCU(A) |
70 |
2.09 |
Asp |
GAU(D) |
51 |
1.52 |
Gly |
GGU(G) |
100 |
2.99 |
Ser |
S |
350 |
10.4 |
|
Val |
GUC(V) |
32 |
0.96 |
Ala |
GCC(A) |
18 |
0.54 |
Asp |
GAC(D) |
15 |
0.45 |
Gly |
GGC(G) |
40 |
1.19 |
Thr |
T |
88 |
2.6 |
|
Val |
GUA(V) |
58 |
1.73 |
Ala |
GCA(A) |
10 |
0.30 |
Glu |
GAA(E) |
15 |
0.45 |
Gly |
GGA(G) |
49 |
1.46 |
Val |
V |
402 |
11.9 |
|
Val |
GUG(V) |
122 |
3.64 |
Ala |
GCG(A) |
29 |
0.87 |
Glu |
GAG(E) |
55 |
1.64 |
Gly |
GGG(G) |
141 |
4.21 |
Trp |
W |
118 |
3.5 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Tyr |
Y |
175 |
5.2 |