Abstract
In order to examine the prevalence of Cryptosporidium infection in wild rodents and insectivores of South Korea and to assess their potential role as a source of human cryptosporidiosis, a total of 199 wild rodents and insectivore specimens were collected from 10 regions of South Korea and screened for Cryptosporidium infection over a period of 2 years (2012-2013). A nested-PCR amplification of Cryptosporidium oocyst wall protein (COWP) gene fragment revealed an overall prevalence of 34.2% (68/199). The sequence analysis of 18S rRNA gene locus of Cryptosporidium was performed from the fecal and cecum samples that tested positive by COWP amplification PCR. As a result, we identified 4 species/genotypes; chipmunk genotype I, cervine genotype I, C. muris, and a new genotype which is closely related to the bear genotype. The new genotype isolated from 12 Apodemus agrarius and 2 Apodemus chejuensis was not previously identified as known species or genotype, and therefore, it is supposed to be a novel genotype. In addition, the host spectrum of Cryptosporidium was extended to A. agrarius and Crosidura lasiura, which had not been reported before. In this study, we found that the Korean wild rodents and insectivores were infected with various Cryptosporidium spp. with large intra-genotypic variationa, indicating that they may function as potential reservoirs transmitting zoonotic Cryptosporidium to livestock and humans.
-
Key words: Cryptosporidium, rodent, insectivore, COWP, 18S rRNA, Korea
Cryptosporidium is an apicomplexan protozoan parasite that infects a wide range of vertebrate species including humans [
1]. This parasite invades the epithelium of the gastrointestinal tract, causing mild-to-severe diarrhea depending on the host’s immune status [
2]. It leads to acute and self-limiting illness when ingested through contaminated water or food in immunocompetent individuals. However, cryptosporidiosis can become a chronic and life-threatening disease in immunocompromised individuals [
3,
4]. Thus, cryptosporidiosis has been a major public health concern at water utilities [
5]. Currently, at least 26 species and 50 different genotypes of
Cryptosporidium have been recognized worldwide through phylogenetic analyses [
6]. Among them,
C. parvum and
C. hominis are widely known as the causative agents of diarrheal illness in humans [
7], although recent studies have suggested that all
Cryptosporidium parasites should be considered hazardous to humans [
8,
9].
Wild mammals, particularly rodents and insectivores, have received attention as important reservoirs of
Cryptosporidium, especially
C. parvum and
C. muris [
10,
11]. Many other species, such as
C. hominis, C. meleagridis, C. felis, C. canis, C. ubiquitum, C. cuniculus, and
C. viatorum, have also been found to infect rodents as well as humans [
6,
8,
12-
14]. Several studies conducted in different countries including Spain, Japan, China, and the Philippines revealed that wild rodents are naturally infected with
Cryptosporidium, and the prevalence rate ranged from 1% to 63% [
1,
6,
10,
15]. These results suggest the potential threat of wild rodents to human health in urban and rural environments. In fact, increased participation in activities, such as camping, hiking or engaging in outdoor sports in natural areas, can result in higher exposure to
Cryptosporidium parasites transmitted through water or wildlife [
16].
Nevertheless, few epidemiological studies have been conducted on
Cryptosporidium in small wild animals in Korea, regardless of the potential risk. Previous studies conducted on humans in South Korea showed
C. parvum infection rates of 0.6-25.9% in healthy subjects and 1.0-5.7% in patients with gastroenteritis [
17-
20]. In livestock, such as cattle and pigs, infection rates were between 9.3% and 94% [
19,
21]. However, these studies were limited, as that they did not perform species/genotype identification which can provide the epidemiological characteristics of
Cryptosporidium and they ignored small animals, such as wild rodents, which can serve as reservoir hosts.
This study was designed to investigate the prevalence of Cryptosporidium in commonly found wild rodents and insectivores in South Korea, to determine their genotypes and evolutionary relationships, and to understand their potential role in transmitting Cryptosporidium. Here, we present the results of a 2-year (2012-2013) investigation of Cryptosporidium infection in wild rodents and insectivores from 10 different regions of South Korea.
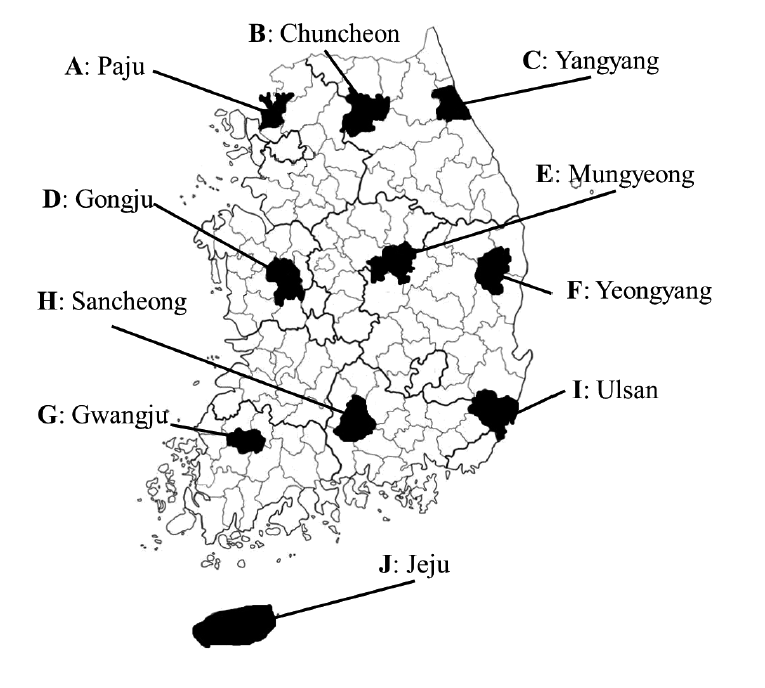
From 2012 to 2013, a total of 199 rodents and insectivores were captured with Sherman traps from 10 regions in South Korea. Each region was designated by the letters from A to J in the order of their documentation (
Fig. 1). They included 169 striped field mice (
Apodemus agrarius), 19 Jeju striped field mice (
Apodemus chejuensis), 10 Ussuri white-toothed shrew (
Crocidura lasiura), and 1 lesser white-toothed shrew (
Crosidura shantungensis).
For molecular detection of
Cryptosporidium from fecal and cecum samples, we have conducted nested-PCR at
Cryptosporidium oocyst wall protein (COWP) gene locus, based on published methods [
22] using 2 primer pairs (primary primers: COWPF1 5´-ACATTTTCAG GAAAGCAGTGTG-3´ and COWPR1 5´-CCTTGCAGTGTGAAATTTGG-3´; nested-primers: COWPF2 5´-CTGATACTGCACCTCCCAAC-3´ and COWPF2 5´-GCTGA TTCAGGTGCCATACA-3´). These primer pairs are optimized to amplify DNA with high sensitivity and without cross-reactivity with other parasite DNA such as that of Eimeria or Cyclospora [
23]. Consequently, the nested-PCR screening detected 68
Cryptosporidium positives from the 199 fecal or cecum DNA samples.
Most of the
Cryptosporidium-positive animals had no apparent clinical signs such as diarrhea or lethargy at the time of sample collection. On the area A through J, the average infection rate of each region was 43.5%, 40.0%, 15.4%, 62.5%, 55.0%, 47.1%, 15.0%, 17.6%, 18.8%, and 31.6%, respectively (
Table 1). The overall prevalence of the infection among rodents and insectivores was 34.2% (68/199) in South Korea, which is within the reported range of 1% to 63% (1, 6, 11, 12, and 32-34). In wild rodents, the prevalence of
Cryptosporidium in
A. agrarius and
A. chejuensis were 33.1% (56/169) and 31.6% (6/19), respectively. These results are similar with previous reports conducted on other wild
Apodemus rodents, which was 35.2% in Spain and 27% in Japan [
10,
15]. In the case of the genus
Crocidura; however, infection rates among
C. lasiura and
C. shantungensis were 60% (6/10) and 0% (0/1), respectively. The ratio showed some difference compared to a previous study which reported the infection rate of 14.8% on C. russula in Spain [
10]. Moreover, there seemed to exist some regional differences in the prevalence of
Cryptosporidium in Korea (
Table 1). Therefore, we have accumulated each data for a long term epidemiological study.
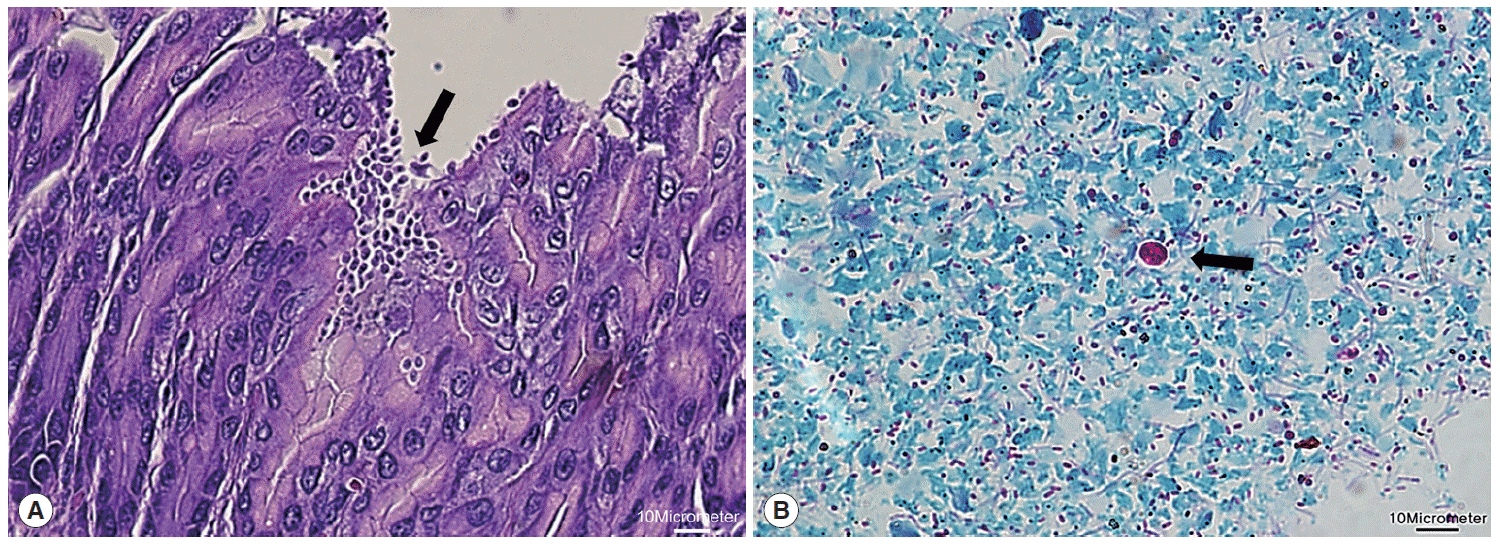
Next, microscopic examinations of H-E stained stomach section taken at necropsy of
Cryptosporidium-positive samples revealed the presence of numerous
Cryptosporidium parasites with eosinophilic small oval structures lining the mucosal gastric epithelia of gastric pits (
Fig. 2A). Moreover, oocysts in fecal samples stained by modified Ziehl-Neelsen technique showed typical characteristics of
Cryptosporidium (
Fig. 2B). The oocyst appeared deep red colored spherical morphology with approximately 4-6 μm in diameter. It contained central black granule and the crescent shaped sporozoites.
Lastly, for the phylogenetic analysis, all samples found to be positive for
Cryptosporidium were also re-amplified at the 18S rRNA gene locus by nested PCR, as described previously [
24], using 2 primer pairs (primary primers: 18SiCF2 5´-GACATATCATTCAAGTTTCTGACC-3´ and 18SiCR2 5´-CTGAAGGAGTAAGGAACAACC-3´; nested primers: 18SiCF1 5´-CCTATCA GCTTTAGACGGTAGG-3´ and 18SiCR1 5´-TCTAAGAATTTCACCTCTGACTG-3´), and sequenced by commercial company (Cosmo-Genetech, Seoul, Korea). The effective length for sequencing was approximately 500 bp because the primer pair (18SiCF1 and 18SiCR1) amplified 587 bp fragments. Of the 68 positive samples identified at the 18S rRNA gene locus, 44 were successful for sequence analysis. The obtained sequences were aligned with reference sequences of
Cryptosporidium species and genotypes from GenBank using ClustalW algorithm of the BioEdit Sequence Alignment Editor software (available at
http://www.mbio.ncsu.edu/bioedit/bioedit.html). The neighbor joining analysis with evolutionary distances calculated using Tamura-Nei parameter model was conducted using MEGA version 5.2 [
25].
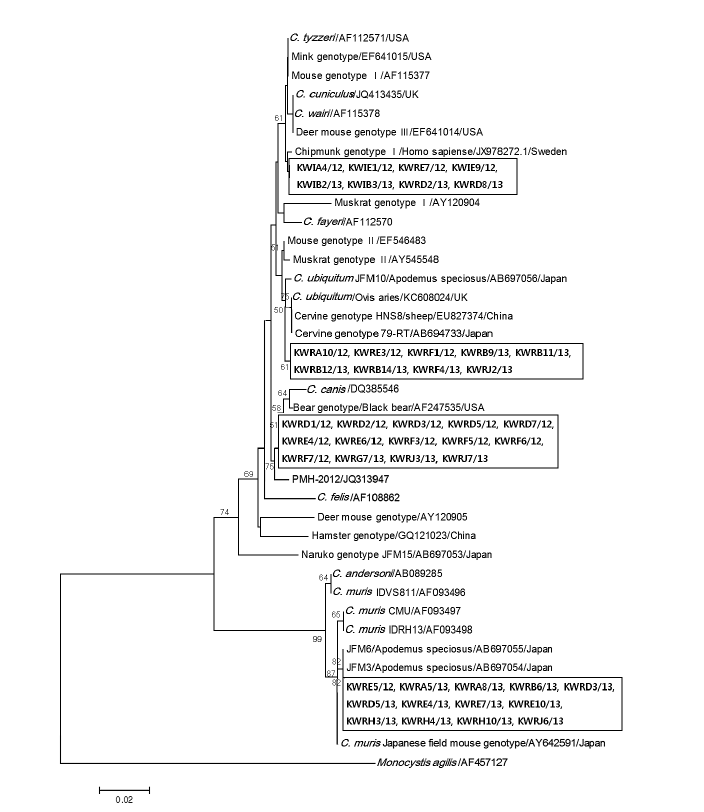
Fig. 3 shows the phylogenetic relationships between various
Cryptosporidium species/genotypes and the 44 isolates from Korean wild rodents and insectivores. The Korean isolates were identified to cluster with 4 different
Cryptosporidium species and genotypes; Chipmunk genotype I (n=8), cervine genotype I (n=9), bear genotype (n=14), and
C. muris (n=13). The differences between reference sequences and new sequence obtained in this study are shown in
Table 2.
Chipmunk genotype I, identified from a total of 8 isolates in this study, was first reported in water after a storm in New York [
26]. This genotype has been identified in wild animals, such as eastern chipmunks, eastern squirrels, and deer mice in New York, as well as red squirrels in Italy [
27,
28]. Cases in which humans were infected with this species have also been reported as well [
7]. In the present study, 3 of the 35 properly sequenced
A. agrarius samples (8.6%) were infected with this genotype. Interestingly, among the 11 captured insectivores, 5 positive subjects infected with
Cryptosporidium species had chipmunk genotype I. The sequences obtained from these isolates had 98.8-99.0% similarity with the reference sequence identified previously in infected Swedes [
29]. This discrepancy is thought to be based on the sequence heterogeneity of the 18S rRNA gene locus [
30]. This is the first report of chipmunk genotype I identified in the genera
Apodemus and
Crocidura, thus expanding its host range.
Cervine genotype I infects humans and various mammals, so they are often called
C. ubiquitum [
31,
32]. After this species was first detected from a pediatric patient in Canada, it was identified in human feces in New Zealand, England, Slovenia, UK, and the US [
7,
33-
37]. Infections with this genotype have occurred in many rodent species such as squirrels, chipmunks, and woodchucks [
28]. According to our survey, 22.9% (8/35) of
A. agrarius and 25% (1/4) of
A. chejuensis were infected with cervine genotype I which was previously reported from surface water in Japan and exhibited 99.2-99.5% sequence similarity. These findings indicate that the Korean isolated
Cryptosporidium is more similar to that from Japan than of other countries such as Spain and the Philippines suggesting a geographical difference in the distribution of the
Cryptosporidium subgenotypes.
The Bear genotype which was first isolated from black bears in the US has not been detected in other hosts [
38]. In the present study, we identified
Cryptosporidium isolates from 12
A. agrarius and 2
A. chejuensis clustered with the bear genotype with a rather low similarity of 92.9-98.6%. These are considered to be distinct new genotypes that have not been reported in other hosts, the unique partial sequence obtained from KWRF1/12 was deposited in the GenBank database under the accession no. KP317127, as a
Cryptosporidium sp. from Korea striped field mouse (KSFM).
C. muris was first recognized in laboratory mice and was also identified in diverse mammals such as mice, rats, Japanese field mice, cats, and cynomolgus monkeys [
28,
39,
40]. In addition,
C. muris has been detected in the feces of an immunosuppressed human with human immunodeficiency virus [
41,
42]. In this study, among the 13 isolates identified as
C. muris,
Cryptosporidium isolated from 8
A. agrarius and 1
A. chejuensis exhibited 100% similarity with
C. muris genotype previously reported from A. speciosus in Japan, and the remaining 4
A. agrarius showed 98.8- 99.8% similarity with intra-genotypic variation [
15].
Our results are the first report verifying Cryptosporidium infection status in wild rodents and insectivores in South Korea. The results revealed high genetic diversity of Cryptosporidium spp. in small wild animals in South Korea. In particular, our results widened the scope of Cryptosporidium hosts. Furthermore, chipmunk genotype I, cervine genotype I, and C. muris identified in this study are known to be able to trigger diseases in livestock and humans when infected. These findings suggest that the rodents and insectivores in South Korea may play a role as potential reservoir hosts that can mediate transmission of Cryptosporidium among them. Continuous surveillance and epidemiological study of Cryptosporidium in biological vectors, particularly rodents and insectivores, are required to prevent the unexpected outbreaks in animals including humans.
Notes
-
We have no conflict of interest related to this work.
This study was partially supported by the Research Institute for Veterinary Science, Seoul National University.
Fig. 1.Ten geographic locations in South Korea where trapping of wild rodents and insectivores was conducted in 2012-2013. Each region was designated by capital letters (A-J).
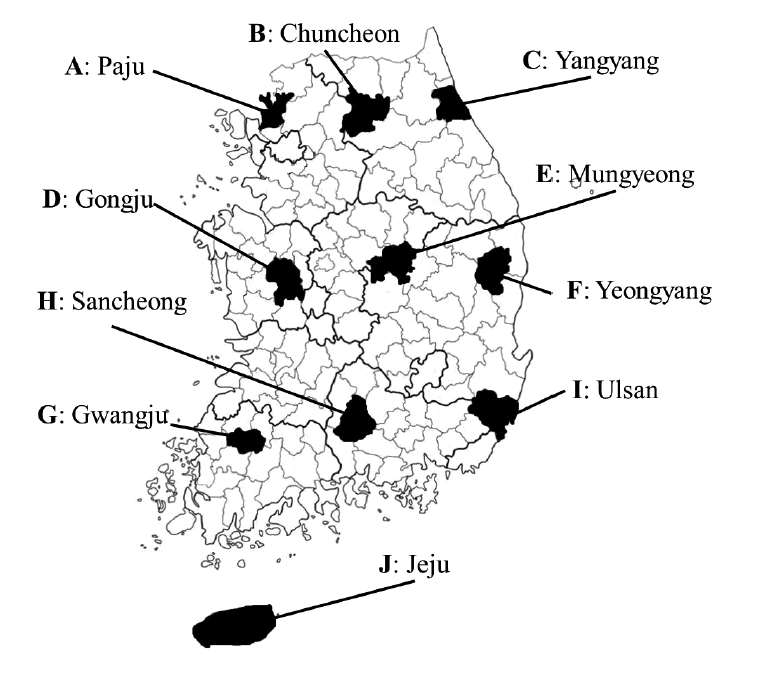
Fig. 2.Histological examination of Cryptosporidium in the stomach. (A) Clusters of Cryptosporidium (arrow) adherent to the luminal part of epithelial cells of stomach villi (H&E stain; ×40). (B) Cryptosporidium oocyst (arrow) showing deep red staining of internal structure (Ziehl-Neelsen stain; ×100). The scale bars represent 10 μm.
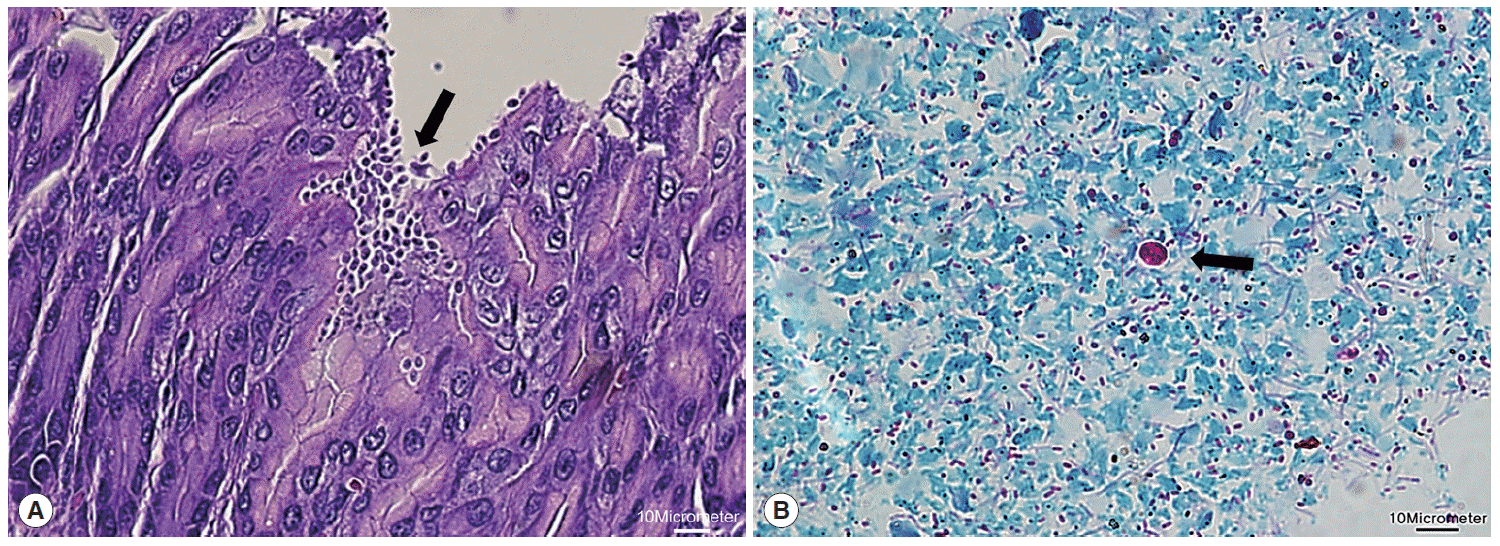
Fig. 3.Evolutionary relationships of Cryptosporidium isolates inferred by a neighbor-joining analysis of Tamura-Nei distances calculated from pairwise comparison of 18S rRNA sequences. The tree was rooted with the 18S rRNA sequence of Monocystis agilis (AF457127). Percentage bootstrap support (>50%) from 1,000 pseudoreplicates is indicated at the left of the supported node. Sample ID indicates: KW (Korean Wild) + R/I (rodent/insectivore) + A-J (Capital letters of each region) + sample number/year of capture.
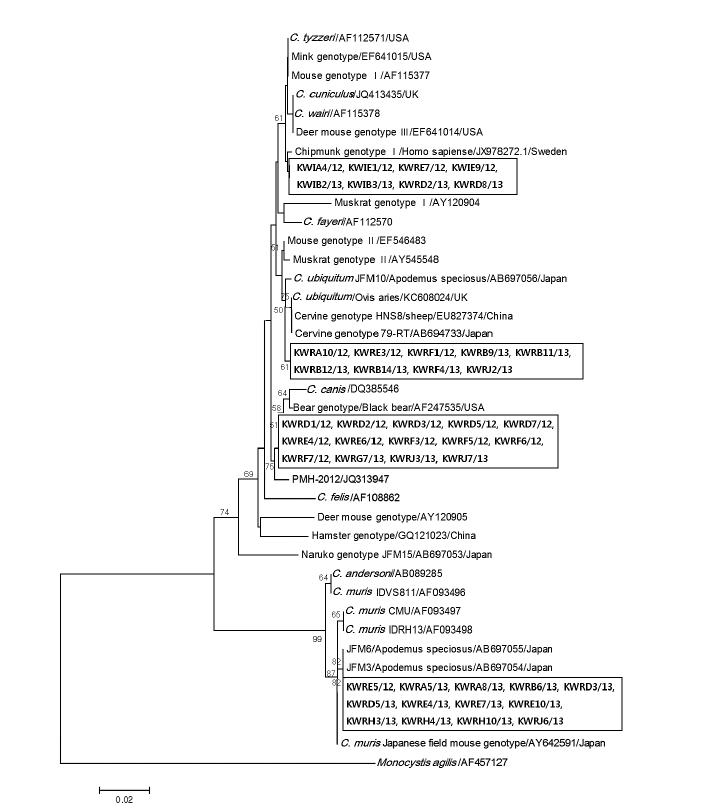
Table 1.Prevalence of detected Cryptosporidium sp. by nested-PCR in wild rodents and insectivores captured in South Korea
Table 1.
|
Location |
Host species |
Total no. of samples |
No. of positive samples |
Prevalence (%) |
Overall prevalence per region (%) |
|
A (paju) |
Apodemus agrarius
|
21 |
8 |
38.1 |
43.5 |
|
Crocidura lasiura
|
2 |
2 |
100.0 |
|
|
B (Chuncheon) |
Apodemus agrarius
|
21 |
8 |
38.1 |
40.0 |
|
Crocidura lasiura
|
4 |
2 |
50.0 |
|
|
C (Yangyang) |
Apodemus agrarius
|
26 |
4 |
15.4 |
15.4 |
|
D (Gongju) |
Apodemus agrarius
|
16 |
10 |
62.5 |
62.5 |
|
E (Mungyeong) |
Apodemus agrarius
|
18 |
9 |
50.0 |
55.0 |
|
Crocidura lasiura
|
2 |
2 |
100.0 |
|
|
F (Yeongyang) |
Apodemus agrarius
|
16 |
8 |
50.0 |
47.1 |
|
Crocidura lasiura
|
1 |
0 |
0.0 |
|
|
G (Gwangju) |
Apodemus agrarius
|
20 |
3 |
15.0 |
15.0 |
|
H (Sancheong) |
Apodemus agrarius
|
15 |
3 |
20.0 |
17.6 |
|
Crocidura lasiura
|
1 |
0 |
0.0 |
|
|
Crocidura shantungensis
|
1 |
0 |
0.0 |
|
|
I (Ulsan) |
Apodemus agrarius
|
16 |
3 |
18.8 |
18.8 |
|
J (Jeju) |
Apodemus chejuensis
|
19 |
6 |
31.6 |
31.6 |
|
Total |
|
199 |
68 |
|
34.2 |
Table 2.Comparison of the nucleotide sequences of the partial 18S rRNA gene of new Cryptosporidium genotype obtained in this study (Cryptosporidium sp. KSFM) and related reference genotypes in the GenBank
Table 2.
|
Organism or genotype (accession no.) |
Nucleotide sequence at the indicated position (s)a
|
|
114-130 |
275-324 |
352-377 |
419-438 |
443-482 |
493-510 |
|
Chipmunk genotype I (jx978272) |

|
|
Cervine genotype I (AB694733) |
|
C. muris (AB697054) |
|
Bear genotype (AF247535) |
|
Cryptosporidium sp. KSFM (KP317127) |
References
- 1. Lv C, Zhang L, Wang R, Jian F, Zhang S, Ning C, Wang H, Feng C, Wang X, Ren X, Qi M, Xiao L. Cryptosporidium spp. in wild, laboratory, and pet rodents in china: prevalence and molecular characterization. Appl Environ Microbiol 2009;75:7692-7699.
- 2. Leitch GJ, He Q. Cryptosporidiosis-an overview. J Biomed Res 2012;25:1-16.
- 3. Kurniawan A, Dwintasari SW, Connelly L, Nichols RA, Yunihastuti E, Karyadi T, Djauzi S. Cryptosporidium species from human immunodeficiency-infected patients with chronic diarrhea in Jakarta, Indonesia. Ann Epidemiol 2013;23:720-723.
- 4. Elwin K, Hadfield SJ, Robinson G, Chalmers RM. The epidemiology of sporadic human infections with unusual cryptosporidia detected during routine typing in England and Wales, 2000-2008. Epidemiol Infect 2012;140:673-683.
- 5. Xiao L, Morgan UM, Fayer R, Thompson RC, Lal AA. Cryptosporidium systematics and implications for public health. Parasitol Today 2000;16:287-292.
- 6. Ng-Hublin JS, Singleton GR, Ryan U. Molecular characterization of Cryptosporidium spp. from wild rats and mice from rural communities in the Philippines. Infect Genet Evol 2013;16:5-12.
- 7. Feltus DC, Giddings CW, Schneck BL, Monson T, Warshauer D, McEvoy JM. Evidence supporting zoonotic transmission of Cryptosporidium spp. in Wisconsin. J Clin Microbiol 2006;44:4303-4308.
- 8. Xiao L, Fayer R, Ryan U, Upton SJ. Cryptosporidium taxonomy: recent advances and implications for public health. Clin Microbiol Rev 2004;17:72-97.
- 9. Chen XM, Keithly JS, Paya CV, LaRusso NF. Cryptosporidiosis. N Engl J Med 2002;346:1723-1731.
- 10. Torres J, Gracenea M, Gomez MS, Arrizabalaga A, Gonzalez-Moreno O. The occurrence of Cryptosporidium parvum and C. muris in wild rodents and insectivores in Spain. Vet Parasitol 2000;92:253-260.
- 11. Kimura A, Edagawa A, Okada K, Takimoto A, Yonesho S, Karanis P. Detection and genotyping of Cryptosporidium from brown rats (Rattus norvegicus) captured in an urban area of Japan. Parasitol Res 2007;100:1417-1420.
- 12. Foo C, Farrell J, Boxell A, Robertson I, Ryan UM. Novel Cryptosporidium genotype in wild Australian mice (Mus domesticus). Appl Environ Microbiol 2007;73:7693-7696.
- 13. Lebbad M, Beser J, Insulander M, Karlsson L, Mattsson JG, Svenungsson B, Axen C. Unusual cryptosporidiosis cases in Swedish patients: extended molecular characterization of Cryptosporidium viatorum and Cryptosporidium chipmunk genotype I. Parasitology 2013;140:1735-1740.
- 14. Robinson G, Elwin K, Chalmers RM. Unusual Cryptosporidium genotypes in human cases of diarrhea. Emerg Infect Dis 2008;14:1800-1802.
- 15. Murakoshi F, Fukuda Y, Matsubara R, Kato Y, Sato R, Sasaki T, Tada C, Nakai Y. Detection and genotyping of Cryptosporidium spp. in large Japanese field mice, Apodemus speciosus. Vet Parasitol 2013;196:184-188.
- 16. Hayes EB. Looking the other way: preventing vector-borne disease among travelers to the United States. Travel Med Infect Dis 2010;8:277-284.
- 17. Cheun HI, Cho SH, Lim YY, Lee BC, Kim JY, Ju JW, Na BK, Kimata I, Yu JR, Kim TS. Cryptosporidium parvum in Korea: prevalence in individuals residing in three major river valleys and genetic characteristics of the isolates. J Vet Med Sci 2010;72:167-172.
- 18. Lee JK, Song HJ, Yu JR. Prevalence of diarrhea caused by Cryptosporidium parvum in non-HIV patients in Jeollanam-do, Korea. Korean J Parasitol 2005;43:111-114.
- 19. Park JH, Guk SM, Han ET, Shin EH, Kim JL, Chai JY. Genotype analysis of Cryptosporidium spp. prevalent in a rural village in Hwasun-gun, Republic of Korea. Korean J Parasitol 2006;44:27-33.
- 20. Huh JW, Moon SG, Lim YH. A survey of intestinal protozoan infections among gastroenteritis patients during a 3-year period (2004-2006) in Gyeonggi-do (province), South Korea. Korean J Parasitol 2009;47:303-305.
- 21. Yu JR, Lee JK, Seo M, Kim SI, Sohn WM, Huh S, Choi HY, Kim TS. Prevalence of cryptosporidiosis among the villagers and domestic animals in several rural areas of Korea. Korean J Parasitol 2004;42:1-6.
- 22. Xiao L, Limor J, Morgan UM, Sulaiman IM, Thompson RC, Lal AA. Sequence differences in the diagnostic target region of the oocyst wall protein gene of Cryptosporidium parasites. Appl Environ Microbiol 2000;66:5499-5502.
- 23. Lee SH, Joung M, Yoon S, Choi K, Park WY, Yu JR. Multiplex PCR detection of waterborne intestinal protozoa: microsporidia, Cyclospora, and Cryptosporidium. Korean J Parasitol 2010;48:297-301.
- 24. Ryan U, Xiao L, Read C, Zhou L, Lal AA, Pavlasek I. Identification of novel Cryptosporidium genotypes from the Czech Republic. Appl Environ Microbiol 2003;69:4302-4307.
- 25. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011;28:2731-2739.
- 26. Jiang J, Alderisio KA, Xiao L. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl Environ Microbiol 2005;71:4446-4454.
- 27. Kvac M, Hofmannova L, Bertolino S, Wauters L, Tosi G, Modry D. Natural infection with two genotypes of Cryptosporidium in red squirrels (Sciurus vulgaris) in Italy. Folia Parasitol (Praha) 2008;55:95-99.
- 28. Feng Y, Alderisio KA, Yang W, Blancero LA, Kuhne WG, Nadareski CA, Reid M, Xiao L. Cryptosporidium genotypes in wildlife from a new york watershed. Appl Environ Microbiol 2007;73:6475-6483.
- 29. Insulander M, Silverlas C, Lebbad M, Karlsson L, Mattsson JG, Svenungsson B. Molecular epidemiology and clinical manifestations of human cryptosporidiosis in Sweden. Epidemiol Infect 2013;141:1009-1020.
- 30. Le Blancq SM, Khramtsov NV, Zamani F, Upton SJ, Wu TW. Ribosomal RNA gene organization in Cryptosporidium parvum . Mol Biochem Parasitol 1997;90:463-478.
- 31. Slapeta J. Cryptosporidiosis and Cryptosporidium species in animals and humans: a thirty colour rainbow? Int J Parasitol 2013;43:957-970.
- 32. Fayer R, Santin M, Macarisin D. Cryptosporidium ubiquitum n. sp. in animals and humans. Vet Parasitol 2010;172:23-32.
- 33. Ong CS, Eisler DL, Alikhani A, Fung VW, Tomblin J, Bowie WR, Isaac-Renton JL. Novel Cryptosporidium genotypes in sporadic cryptosporidiosis cases: first report of human infections with a cervine genotype. Emerg Infect Dis 2002;8:263-268.
- 34. Learmonth JJ, Ionas G, Ebbett KA, Kwan ES. Genetic characterization and transmission cycles of Cryptosporidium species isolated from humans in New Zealand. Appl Environ Microbiol 2004;70:3973-3978.
- 35. Leoni F, Amar C, Nichols G, Pedraza-Diaz S, McLauchlin J. Genetic analysis of Cryptosporidium from 2414 humans with diarrhoea in England between 1985 and 2000. J Med Microbiol 2006;55:703-707.
- 36. Soba B, Petrovec M, Mioc V, Logar J. Molecular characterisation of Cryptosporidium isolates from humans in Slovenia. Clin Microbiol Infect 2006;12:918-921.
- 37. Davies AP, Campbell B, Evans MR, Bone A, Roche A, Chalmers RM. Asymptomatic carriage of protozoan parasites in children in day care centers in the United kingdom. Pediatr Infect Dis J 2009;28:838-840.
- 38. Xiao L, Limor JR, Sulaiman IM, Duncan RB, Lal AA. Molecular characterization of a Cryptosporidium isolate from a black bear. J Parasitol 2000;86:1166-1170.
- 39. Iseki M, Maekawa T, Moriya K, Uni S, Takada S. Infectivity of Cryptosporidium muris (strain RN 66) in various laboratory animals. Parasitol Res 1989;75:218-222.
- 40. Feng Y. Cryptosporidium in wild placental mammals. Exp Parasitol 2010;124:128-137.
- 41. Palmer CJ, Xiao L, Terashima A, Guerra H, Gotuzzo E, Saldias G, Bonilla JA, Zhou L, Lindquist A, Upton SJ. Cryptosporidium muris, a rodent pathogen, recovered from a human in Peru. Emerg Infect Dis 2003;9:1174-1176.
- 42. Gatei W, Ashford RW, Beeching NJ, Kamwati SK, Greensill J, Hart CA. Cryptosporidium muris infection in an HIV-infected adult, Kenya. Emerg Infect Dis 2002;8:204-206.