Abstract
Echinococcus granulosus sensu lato (s.l.) is a causative agent of cystic echinococcosis or cystic hydatid disease in humans and domestic and wild animals. The disease is a serious health problem in countries associated with poverty and poor hygiene practices, particularly in livestock raising. We introduced a practical algorism for genotyping the parasite, which may be useful to many developing countries. To evaluate the efficiency of the algorism, we genotyped 3 unknown strains isolated from human patients. We found that unknowns 1 and 3 were included in G1, G2, and G3 genotypes group and unknown 2 was included in G4 genotype (Echinococcus equinus) according to the algorisms. We confirmed these results by sequencing the 3 unknown isolates cox1 and nad1 PCR products. In conclusion, these new algorisms are very fast genotype identification tools that are suitable for evaluating E. granulosus s.l. isolated from livestock or livestock holders, particularly in developing countries.
-
Key words: Echinococcus granulosus, practical algorism, genotyping, cox1, nad1
Echinococcus granulosus sensu lato (s.l.) is a causative agent of cystic echinococcosis or cystic hydatid disease in humans and domestic and wild animals [
1]. This disease is a serious health problem in countries associated with poverty and poor hygiene practices, particularly livestock raising [
2]. Until recently, the taxonomy of
E. granulosus s.l. spp. has been controversial because of the limited availability of morphological diagnostic characters in both adult and larval stages [
3]. The incomplete development of metacestodes in aberrant hosts has further limited the identification of causative organisms. In the 1990s, an Australian research group [
4–
6] conducted molecular taxonomic analyses of
E. granulosus s.l. using short mitochondrial DNA sequences of genes for cytochrome
c oxidase subunit 1 (
cox1) and NADH dehydrogenase subunit 1 (
nad1). The analyses showed that
Echinococcus multilocularis, Echinococcus vogeli, and
Echinococcus oligarthra are distinct from each other and that
E. granulosus s.l. can be divided into genotypes G1–G8 [
7]. These genotypes correspond to the following strains:
E. granulosus sensu stricto (s.s.) G1 (sheep strain), G2 (Tasmanian sheep strain), G3 (buffalo strain);
Echinococcus equinus G4 (horse strain);
Echinococcus ortleppi G5 (cattle strain);
Echinococcus canadensis G6 (camel strain), G7 (pig strain), and G8 (cervid strain) [
3,
8]. Currently, these genotypes are used as standards for molecular classification when considering the epidemiology of cystic echinococcosis [
9]. Another
E. canadensis G9 (unidentified strain) and G10 (Fennoscandian cervid strain) were subsequently characterized [
10,
11], while the status of G9 remains unclear [
3,
8].
Although entire or partial gene sequencing is effective for determining genotypes, specialized equipment is required to conduct these costly and time-consuming procedures. In many developing countries, the supply of such equipment is insufficient, and thus PCR-RFLP is frequently used [
12,
13]. In fact, most countries in which this parasite is prevalent are economically underdeveloped, and thus a simple PCR-RFLP method is necessary. However, PCR-RFLP results must be compared with previously reported PCR-RFLP results. Using this method, some under-digested DNA fragments may be confused with fully digested fragments, limiting the interpretation of PCR-RFLP results. To overcome this limitation, we developed a PCR-RFLP algorism involving only 2 or 3 restriction enzymes that produce only 1 or 2 fragments in PCR-RFLP, which is highly helpful for improving the quality of the results.
The
cox1 and
nad1 sequences from 9
E. granulosus s.l. genotypes (G1–G10, except G9) were downloaded from GenBank (
https://www.ncbi.nlm.nih.gov/nucgss). After multiple alignment (MEGA software version 6), we searched for new PCR target lesions that could produce 1 or 2 fragments after restriction endonuclease digestion using NEBcutter version 2.0 (
http://nc2.neb.com/NEBcutter2/). The PCR primer sets for
cox1 and
nad1 are constructed like as cox-f [CC(C/T) GG(A/G) TTT GGT(A/G) T(A/T) ATT], cox-r [ATC(A/G) TG(C/T) AA AA(C/T) ATT ATC], nad-f [A(G/A)(A/T) TTC GTA AGG G(G/C) C CTA ATA], and nad-r [(A/T)CC(A/T) CT AAC(T/C) AA TTC ACT TTC]. We predicted hypothetical PCR-RFLP fragments based on various endonuclease recognition sites, and selected appropriate restriction enzyme sets for each
cox1 and
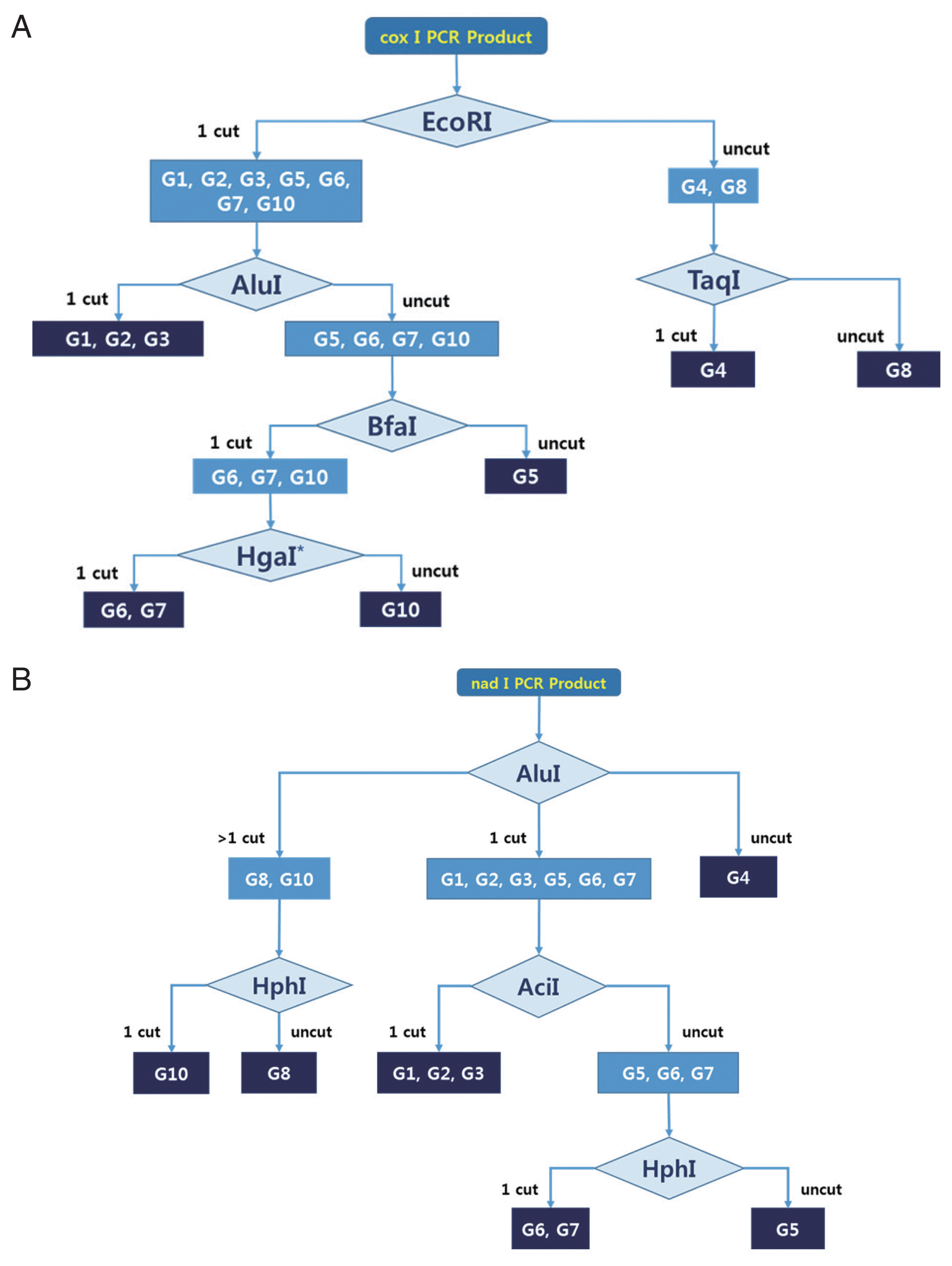
nad1 PCR RFLP algorism. We used more than 10 restriction enzymes for hypothetical RFLP, as it was not possible to identify all genotypes using only 1 enzyme. Our goal was to develop an algorism that could identify all genotypes using the fewest enzymes. Most genotypes could be distinguished by using a specific restriction enzyme (
Supplementary Fig. 1). However, we could not distinguish between the G1–G2, G1–G3, G2–G3, and G6–G7 genotypes using
cox1 and
nad1 PCR-RFLP in this study. Finally, we determined the best algorism for 2 mitochondrial genes (
cox1, nad1) by comparing band patterns (
Fig. 1). To evaluate the algorism, unidentified genomic DNA was isolated from 3
Echinococcus protoscoleces. Three hydatid cyst fluids were collected from patients who underwent medical treatment at Samarqand Hospital in Uzbekistan. The protoscoleces were obtained after centrifugation for 5 min at 2,000 ×g. Total genomic DNA was extracted using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer instructions. The genomic DNA was eluted in 200 μl of water and stored at −20°C until PCR amplification. The extracted genomic DNA was amplified using noble primer sets for 2 mitochondrial DNA genes. PCR was carried out in a final volume of 50 μl containing 5 μl genomic DNA, 10X EF-Taq Reaction Buffer (25 mM MgCl
2 mixed), 2 μl of 10 mM dNTPs, 5 μl of 10 pmol of each primer, and 1 μl (2.5 U) of DiaStar
TM EF-Taq DNA Polymerase in a thermocycler (Takara, Shiga, Japan).
The following temperature profile was used for
cox1 amplification: initial denaturation at 94°C for 30 sec; followed by 35 cycles of denaturation at 94°C for 30 sec, annealing at 42°C for 45 sec, and elongation at 72°C for 35 sec. This was followed by final extension at 72°C for 10 min. The following temperature profile was used for
nad1 amplification: initial denaturation at 94°C for 30 sec; followed by 35 cycles of denaturation at 94°C for 30 sec, annealing at 45°C for 45 sec, and elongation at 72°C for 35 sec. Final extension was performed at 72°C for 10 min. PCR products were confirmed on 1.0% agarose gels under ultraviolet (UV) light. The PCR products were digested for 2–3 hr at 37°C by 4 restriction enzymes (
AluI,
EcoRI,
AciI, and
HphI) and at 65°C by the restriction enzyme
TaqI. Restriction fragments were separated on 1.5% agarose gels and photographed under UV light. After 1% agarose gel electrophoresis of
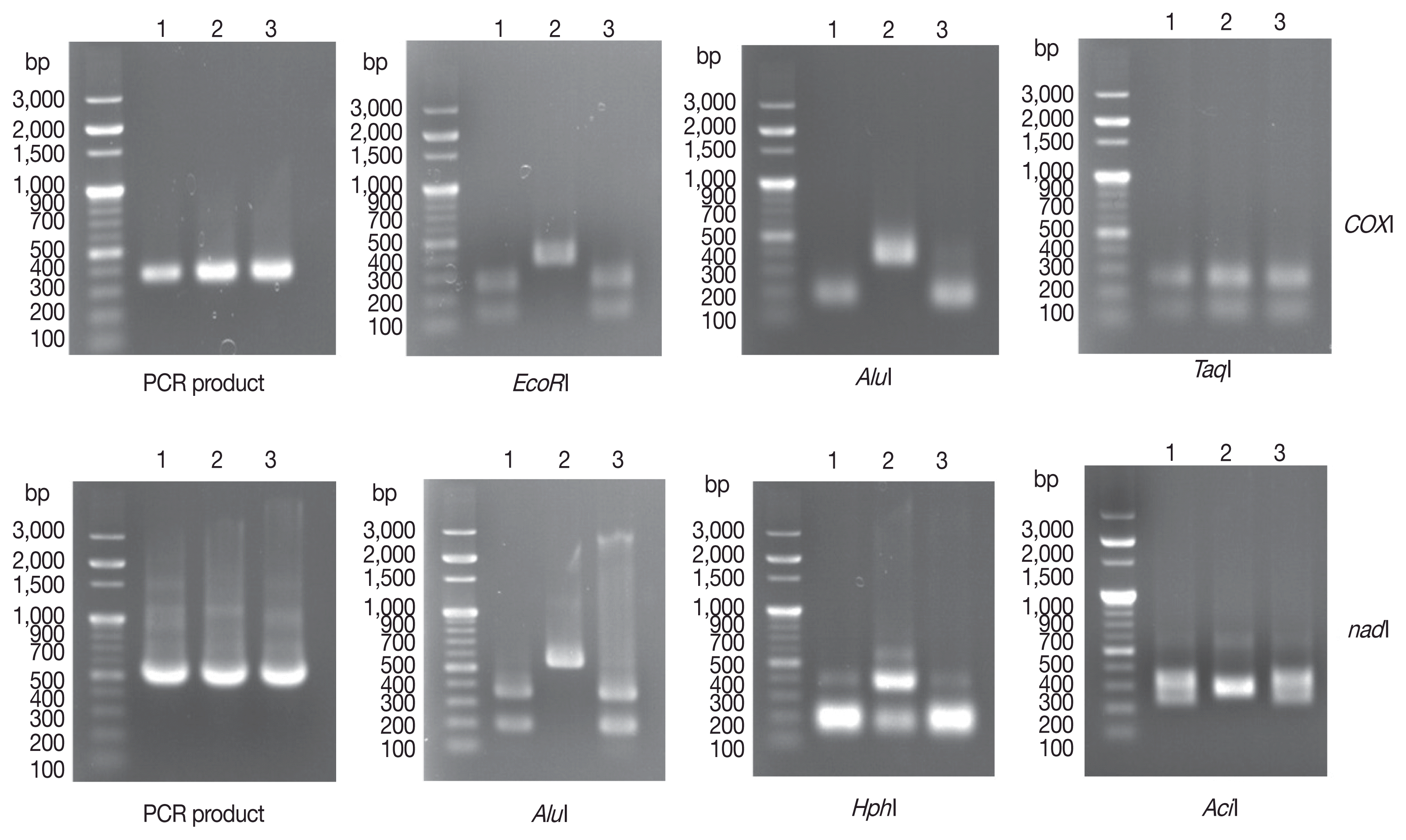
cox1 and
nad1 PCR products, we obtained 350-base pair (bp) (expected size 366 bp)
cox1 PCR products and 500-bp (expected size 530 bp)
nad1 PCR products (
Fig. 2) from 3 human
Echinococcus sample. After digestion of
cox1 gene PCR products with
EcoRI, 2 PCR-RFLP patterns were observed. The genotypes of the 2 samples (unknowns 1 and 3) were likely G1, G2, G3, G5, G6, G7, or G10, while that of unknown 2 was likely G4 or G8. After
AluI digestion, we found that unknowns 1 and 3 were included in
E. granulosus s.s (G1, G2, and G3 genotypes). Finally, we found that unknown 2 was included in
E. equinus (G4 genotype) based on
TaqI digestion. For the
nad1 PCR-RFLP genotyping algorism, the same results were observed for 3 different restriction enzymes (
AluI,
HphI, and
AciI) (
Fig. 2).
To confirm the PCR-RFLP algorism,
nad1 and
cox1 PCR products of unidentified genotypes were sequenced after purifying the PCR products with Expin
TM PCR SV (GeneAll Biotech, Seoul, Korea). Sequences and the reference
E. granulosus s.l. genotype sequences were subjected to multiple alignment and phylogenetic distances were calculated using BLAST (
http://www.ncbi.nlm.nih.gov/) and MEGA software version 6. Phylogenetic trees were constructed using the neighbor-joining method in MEGA software version 6.0. DNA sequencing yielded consensus sequences of the
cox1 and
nad1 genes. Key reference sequences for genotyping were used according to previous studies [
14,
15]. Sequences analysis of
cox1 PCR products using alignment and phylogenetic analysis with reference sequences indicated that unknowns 1 and 3 were very similar to the sequences of G1 (99% identity), G2 (99% identity) and G3 (99% identity). The entire sequence of
cox1 PCR product of unknown 2 was identical to that of the G4 genotype (
Fig. 3). For
nad1 gene sequence analysis, although there were some insignificant differences, we obtained similar results as those obtained for
cox1 (
Fig. 3).
Based on morphological characteristics, 4 species are currently recognized in the genus
Echinococcus: E. multilocularis, E. vogeli, E. oligarthra, and
E. granulosus s.l. Other species such as
E. shiquicus, E. felidis, E. ortleppi, E. equinus, E. canadensis and
E. intermedius are currently classified as
E. granulosus s.l. [
3,
8,
16,
17]. We constructed practical algorism for rapid genotyping of
E. granulosus s.l. isolated from human and domestic animals using only 1–3 restriction enzymes. This method is less cost and more rapid than sequencing analysis. Although G1, G2, and G3 were not distinguished by this algorism, these genotypes have been recognized as the same species,
E. granulosus s.s. [
15,
18,
19]. The algorism distinguished G8 and G10 from the G6/G7 complex, but
E. canadensis G6 and G7 could not be distinguished. Previous studies were also unable to separate the G6/G7 complex using mitochondria DNA, ITS1, and 18S ribosomal DNA sequencing [
8,
20].
There were some limitations to this study. The algorisms were developed for only 9 genotypes (G1–G10, except G9) of
E. granulosus s.l. and not for
E. felidis, E. multilocularis, E. shiquicus, E. oligarthra, and
E. vogeli, which use wild animals as their intermediated hosts [
8]. However, among these species, only
E. multilocularis is known to commonly infect humans and is not found in domestic animals. Additionally, the morphological characteristics of their hydatid cysts are quite different from those of
E. granulosus s.l. [
20]. In these cases, genetic classification may be unnecessary for specific identification. Interestingly, we identified the first human infection case of
E. equinus (
Fig. 3). No human infections by the G4 genotype have been detected previously [
21].
Supplementary materials
Supplementary Fig. 1
Schematic representation of hypothetical RFLP of E. granulosus s.l. species using four restriction enzymes. (A) The hypothetical pattern of bands using PCR product of coxI by AluI, EcoRI, TaqI, BfaI, and HgaI. (B) The hypothetical pattern of bands using PCR products of and 1 gene by AluI, HphI, and AciI. The 100 bp DNA Ladder Marker (Enzynomics, Daejeon, Korea) was used as a size marker.
Notes
-
CONFLICT OF INTEREST
We have no conflict of interest related to this study.
ACKNOWLEDGMENT
This work was supported by the Korea International Cooperation Agency (KOICA) under the title of “Capacity Building of Infectious and Parasitic Diseases Control in the Republic of Uzbekistan” in 2014 (no. P2014-00076-1).
Fig. 1A practical algorism for E. granulosus s.l. genotyping using PCR-RFLP of cox1 and nad1. Schematic representation of hypothetical RFLP by several restriction enzymes. (A) Algorism for cox1 PCR-RFLP genotyping. HgaI* could be replaced with Tsp45I or BsaHI. (B) Algorism for nad1 PCR-RFLP genotyping.

Fig. 2Agarose gel electrophoretogram of PCR products and PCR-RFLP of 3 unidentified Echinococcus sp. isolated from echinococcosis patients. After PCR, each 5 μl PCR products of cox1 and nad1 were loaded onto a 1.0% agarose gel with 100-bp DNA ladder. The cox1 gene PCR products were digested with EcoRI, AluI, and TaqI according to noble genotyping algorism. For nad1 PCR products, AluI, HphI, and AciI were used for PCR-RFLP algorism. No. 1 and No. 3 isolates were identified as E. granulosus s.s. (G1–G3), but No 2. isolate was identified as E. equinus (G4). The 100-bp DNA Ladder Marker (Enzynomics) was used as a size marker.

Fig. 3Phylogenetic relationship of isolates and other genotypes previously identified. The cox1 and nad1 PCR products of E. granulosus isolates were sequenced and aligned with reference sequences.

References
- 1. Thompson RCA, Lymbery AJ.
Echinococcus and Hydatid Disease. Wallingford, Oxfordshire, UK. CAB International; 1995, p xii, 477.
- 2. Cardona GA, Carmena D. A review of the global prevalence, molecular epidemiology and economics of cystic echinococcosis in production animals. Vet Parasitol 2013;192:10-32.
- 3. Knapp J, Gottstein B, Saarma U, Millon L. Taxonomy, phylogeny and molecular epidemiology of Echinococcus multilocularis: from fundamental knowledge to health ecology. Vet Parasitol 2015;213:85-91.
- 4. Bowles J, Blair D, McManus DP. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol Biochem Parasitol 1992;54:165-173.
- 5. Bowles J, McManus DP. Rapid discrimination of Echinococcus species and strains using a polymerase chain reaction-based RFLP method. Mol Biochem Parasitol 1993;57:231-239.
- 6. Bowles J, McManus DP. Molecular variation in Echinococcus
. Acta Trop 1993;53:291-305.
- 7. Bowles J, McManus DP. NADH dehydrogenase 1 gene sequences compared for species and strains of the genus Echinococcus
. Int J Parasitol 1993;23:969-972.
- 8. Nakao M, Lavikainen A, Yanagida T, Ito A. Phylogenetic systematics of the genus Echinococcus (Cestoda: Taeniidae). Int J Parasitol 2013;43:1017-1029.
- 9. McManus DP, Thompson RC. Molecular epidemiology of cystic echinococcosis. Parasitology 2003;127(suppl):37-51.
- 10. Scott JC, Stefaniak J, Pawlowski ZS, McManus DP. Molecular genetic analysis of human cystic hydatid cases from Poland: identification of a new genotypic group (G9) of Echinococcus granulosus
. Parasitology 1997;114:37-43.
- 11. Lavikainen A, Lehtinen MJ, Meri T, Hirvelä-Koski V, Meri S. Molecular genetic characterization of the Fennoscandian cervid strain, a new genotypic group (G10) of Echinococcus granulosus
. Parasitology 2003;127:207-215.
- 12. Chaabane-Banaoues R, Oudni-M’rad M, M’Rad S, Amani H, Mezhoud H, Babba H. A novel PCR-RFLP assay for molecular characterization of Echinococcus granulosus sensu lato and closely related species in developing countries. Parasitol Res 2016;115:3817-3824.
- 13. Moghaddas E, Borji H, Naghibi A, Shayan P, Razmi GR. Molecular genotyping of Echinococcus granulosus from dromedaries (Camelus dromedarius) in eastern Iran. J Helminthol 2015;89:100-104.
- 14. Ahmed ME, Eltom KH, Musa NO, Ali IA, Elamin FM, Grobusch MP, Aradaib IE. First report on circulation of Echinococcus ortleppi in the one humped camel (Camelus dromedaries), Sudan. BMC Vet Res 2013;9:127.
- 15. Sharbatkhori M, Fasihi Harandi M, Mirhendi H, Hajialilo E, Kia EB. Sequence analysis of cox1 and nad1 genes in Echinococcus granulosus G3 genotype in camels (Camelus dromedarius) from central Iran. Parasitol Res 2011;108:521-527.
- 16. Thompson RC, McManus DP. Towards a taxonomic revision of the genus Echinococcus
. Trends Parasitol 2002;18:452-457.
- 17. Thompson RC, Lymbery AJ, Constantine CC. Variation in Echinococcus: towards a taxonomic revision of the genus. Adv Parasitol 1995;35:145-176.
- 18. Piccoli L, Bazzocchi C, Brunetti E, Mihailescu P, Bandi C, Mastalier B, Cordos I, Beuran M, Popa LG, Meroni V, Genco F, Cretu C. Molecular characterization of Echinococcus granulosus in southeastern Romania: evidence of G1–G3 and G6–G10 complexes in humans. Clin Microbiol Infect 2013;19:578-582.
- 19. Boubaker G, Macchiaroli N, Prada L, Cucher MA, Rosenzvit MC, Ziadinov I, Deplazes P, Saarma U, Babba H, Gottstein B, Spiliotis M. A multiplex PCR for the simultaneous detection and genotyping of the Echinococcus granulosus complex. PLoS Negl Trop Dis 2013;7:e2017.
- 20. Nakao M, McManus DP, Schantz PM, Craig PS, Ito A. A molecular phylogeny of the genus Echinococcus inferred from complete mitochondrial genomes. Parasitology 2007;134:713-722.
- 21. Alvarez Rojas CA, Romig T, Lightowlers MW.
Echinococcus granulosus sensu lato genotypes infecting humans--review of current knowledge. Int J Parasitol 2014;44:9-18.