Abstract
The pathogenesis of cerebral malaria is biologically complex and involves multi-factorial mechanisms such as microvascular congestion, immunopathology by the pro-inflammatory cytokine and endothelial dysfunction. Recent data have suggested that a pleiotropic T-cell immunomodulatory protein (TIP) could effectively mediate inflammatory cytokines of mammalian immune response against acute graft-versus-host disease in animal models. In this study, we identified a conserved homologue of TIP in Plasmodium berghei (PbTIP) as a membrane protein in Plasmodium asexual stage. Compared with PBS control group, the pathology of experimental cerebral malaria (ECM) in rPbTIP intravenous injection (i.v.) group was alleviated by the downregulation of pro-inflammatory responses, and rPbTIP i.v. group elicited an expansion of regulatory T-cell response. Therefore, rPbTIP i.v. group displayed less severe brain pathology and feverish mice in rPbTIP i.v. group died from ECM. This study suggested that PbTIP may be a novel promising target to alleviate the severity of ECM.
-
Key words: Plasmodium berghei, T-cell immunomodulatory protein, inflammatory cytokine, experimental, cerebral malaria, immunopathology
INTRODUCTION
Malaria is caused by protozoan
Plasmodium spp., and is considered as a serious public-health problem in the developing countries.
Plasmodium infection attributed to 200 million new cases and 435,000 deaths globally in 2017 [
1]. Cerebral malaria (CM) is the most severe neurological complication of
Plasmodium falciparum infections [
2], and 10–20% of the survival cases have neurological disabilities [
3,
4].
CM is a severe clinical syndrome and involves multiple pathophysiological processes. Accumulated evidences indicated that an imbalance in pro- and anti-inflammatory immune responses to
Plasmodium infection, disruption of blood-brain barrier (BBB) and endothelial cell activation are central pathophysiological processes in the development of CM [
5]. The pathogenesis of CM likely involves excess production of pro-inflammatory cytokines such as tumor necrosis factor-alpha (TNF-α), gamma interferon (IFN-γ), interleukin-1 (IL-1), and IL-12 [
6–
8]. The up-regulated expression of pro-inflammatory cytokines induces adhesion molecules such as ICAM-1, VCAM-1 and CD36 in brain endothelial cells. It is characterized by cytoadherence of pRBCs, platelets and leukocytes to the brain endothelium, which correspondingly obstructs the brain micro-vessels, leading to the disruption of BBB and pathologic damage of brain micro-vessels [
9,
10].
TIP is a pleiotropic T-cell immunomodulatory protein that plays a crucial role in the mammalian immune responses. Fiscella et al. [
11] reported that the levels of inflammatory cytokines such as IFN-γ, TNF-α and IL-10 were effectively regulated by TIP, inducing protection against acute graft-versus-host disease. In addition, Nono et al. [
12] also confirmed that EmTIP is an
Echinococcus multilocularis homologue of human TIP, and exists during the early stage of
E. multilocularis. It has immunomodulatory effects on CD4
+ T-cells and promotes the production of IFN-γ by regulating Th1 immune response during the early stages of infection. In this study, PbTIP was identified and the protecting effect against ECM of PbTIP was assessed.
MATERIALS AND METHODS
Animals and parasites
Female C57BL/6 and BALB/c mice (6 to 8 weeks old) were purchased from Beijing Animal Institute (Beijing, China). All experimental animals were implemented according to the Animal Care Institute of China Medical University and complied with the Institution Committee-approved protocols.
P. berghei (ANKA strain 2.34) was maintained by serial mechanical passages and prepared for challenge infection [
13].
For the expression of PbTIP, a segment of it encoding 204–335 amino acids was amplified from P. berghei genomic DNA with forward primer (3′-sequence-5′) and reverse primer (3′-sequence-5′). Using restriction enzymes BamHI and XhoI, rPbTIP was cloned into the expression vector pET32a (+) (Novagen, Darmstadt, Germany). The recombinant plasmid was transformed into Escherichia coli BL-21 (Novagen, Darmstadt, Germany). His-tagged rPbTIP was expressed at 20°C for 12 hr with 1 mmol isopropyl-β-D-thiogalactopyranoside (Sigma-Aldrich, St. Louis, Missouri, USA). Ultrafiltration was performed to remove the endotoxin from the bacterially produced protein (endotoxin<1 Eu/μg). The soluble rPbTIP was purified by Ni-NTA His-Bind Superflow (Invitrogen, Carlsbad, California, USA) in accordance to the manufacturer’s instructions. Purified rPbTIP was dialyzed with phosphate buffered saline (PBS) for 4 hr at 4°C. Then the purified recombinant PbTIP was analyzed on 10% SDS-PAGE gel.
Immunization and polyclonal antibody production
To obtain polyclonal anti-rPbTIP antisera, 10 female BALB/c mice were immunized with rPbTIP (50 μg/mouse) using complete Freund’s adjuvant (Sigma-Aldrich). Mice were given booster immunizations of rPbTIP (25 μg/mouse) twice at 3-week intervals with incomplete Freund’s adjuvant. In the control group, mice were immunized with PBS in adjuvant formulations. After immunization, blood was collected via cardiac puncture from mice under anesthesia. Immune responses to antisera of rPbTIP was detected by ELISA as previously described [
14].
After 4 days post-infection (p.i.), blood-stage parasites from the infected mice were cultured using blood-stage culture medium with the following: RPMI 1640, 25% (v/v) fetal calf serum (FCS), 50 mg/L penicillin and streptomycin at 37°C overnight [
15]. On the following day, the culture was fractionated on 55% (v/v) Nycodenz-RPMI (Axis-Shield Poc, Olso, Norway). The gray layer of the interface consisted of concentrated schizonts, which was then collected and washed with PBS. On day 4 of p.i., mice were treated with sulfadiazine (Sigma, 20 mg/L, drinking water) for 2 days to collect the gametocytes. Blood was then purified on 48% (v/v) Nycodenz-RPMI, and the interface layer was collected and washed [
16]. Parasitaemia was allowed to reach 1–3% on day 3 of p.i. for ookinetes purification. Collected blood was diluted to 1:10 with ookinetes medium (RPMI 1640, 25% [v/v] FCS, 50 mg/L penicillin and streptomycin at pH 8.3) at 19°C for 24 hr. Then the ookinetes were separated on 62% (v/v) Nycodenz-RPMI. In order to obtain the lysate, the purified parasites were diluted in 5 ml of 0.17 mol NH
4Cl for 10 min on ice to lyse the iRBCs. Then the cytoplasmic, nuclear and membrane proteins of PbTIP from separated schizonts, gametocytes and ookinetes were extracted using Minute
™ Plasma Membrane Protein Isolation and Cell Fractionation Kit (Invent Biotechologies, Plymouth, Minnesota, USA) according to the manufacturer’s instructions.
The isolated Plasmodium protein (cytoplasmic, nuclear and membrane protein), with 30 μg of 3 representative stages (schizonts, gametocytes and ookinetes) were electrophoresed on 10% SDS-PAGE gel and transferred on to 0.22 μm PVDF membranes (Bio-Rad, Hercules, California, USA). Membranes were then incubated with 5% non-fat milk with TBST for 2 hr and incubated with anti-rPbTIP antisera (1:500). After washing 3 times with TBST, the membranes were incubated with HRP-conjugated goat-anti-mouse antibodies. Proteins on the blot were visualized with a Pierce ECL Western blotting kit (Thermo Scientific, Waltham, Massachusetts, USA).
Indirect Immunofluorescence assay (IFA)
Purified parasites were fixed with 4% paraformaldehyde (Sigma-Aldrich) in PBS for 30 min at room temperature [
17]. Slides were incubated with anti-rPbTIP antisera (1:100) for 1 hr at 37°C. The slides were then washed twice with PBS and incubated with FITC-conjugated goat anti-mouse antibody (1:500) (Invitrogen) for 30 min at 37°C. Nuclei were stained with 4, 6-diamidino-2-phenylindole (DAPI; Invitrogen) 1 μg/ml. Stained parasites were mounted with ProLong® Gold antifade reagent (Invitrogen), and captured on fluorescence microscopy (Olympus, Tokyo, Japan).
Forty female C57BL/6 mice were infected with intraperitoneal (i.p.) injection of 5×10
6 WT
P. berghei ANKA pRBCs [
18]. Then the experimental mice were divided into 2 groups of 20 each: PBS i.v. injection group on days 1, 0, 1, 3, 5, and 7; rPbTIP i.v. injection group 2 mg/kg on days 1, 0, 1, 3, 5, and 7 [
11]. The percentage parasitaemia of Giemsa-stained thin smears from tail blood were monitored by light microscopy and determined by the formula (the number of pRBCs/total number of RBCs)×100. ECM symptoms evaluated were as follows: ruffled fur, respiratory distress, limb paralysis, wobbly gait, hunching, paroxysmal shivering and coma [
19]. When the ECM symptoms were performed, BBB was evaluated by i.v. injection using 200 μl of 2% (wt/vol) Evans blue solution (Sigma-Aldrich) at 6 days p.i. of 3 mice in each group. After 1 hr, the mice were euthanized and perfused with 20 ml of 0.9% NaCl. Brains were taken out and immersed in 2 ml of formamide for 48 hr at 37°C to extract Evans Blue, which was determined by measuring the absorbance at 620 nm [
20,
21]. When the mice were evaluated for neurological symptoms on day 6, brains of 5 mice from each group were removed, fixed in 4% paraformaldehyde for 24 hr and immersed into paraffin. Serial 4 μm thick brain sections were made and stained with hematoxylin and eosin (HE) to examine for micro-vascular obstruction and leakage. Along the endothelial lining, immunohistochemical staining was detected with specific polyclonal antibodies against ICAM-1 (Abcam, Cambridge, UK), VCAM-1 (Santa Cruz Biotechnology, Santa Cruz, California, USA) and CD36 (Santa Cruz Biotechnology) as previously described [
22,
23]. Followed by incubation with biotin-conjugated secondary antibodies, and streptavidin-conjugated peroxidase and the color development was shown using 3-amino-9-ethylcarbazole as the substrate. Nuclei were counterstained with hematoxylin. Positive vessels were visualized by microscopy at ×400 and counted in 20 fields per mouse.
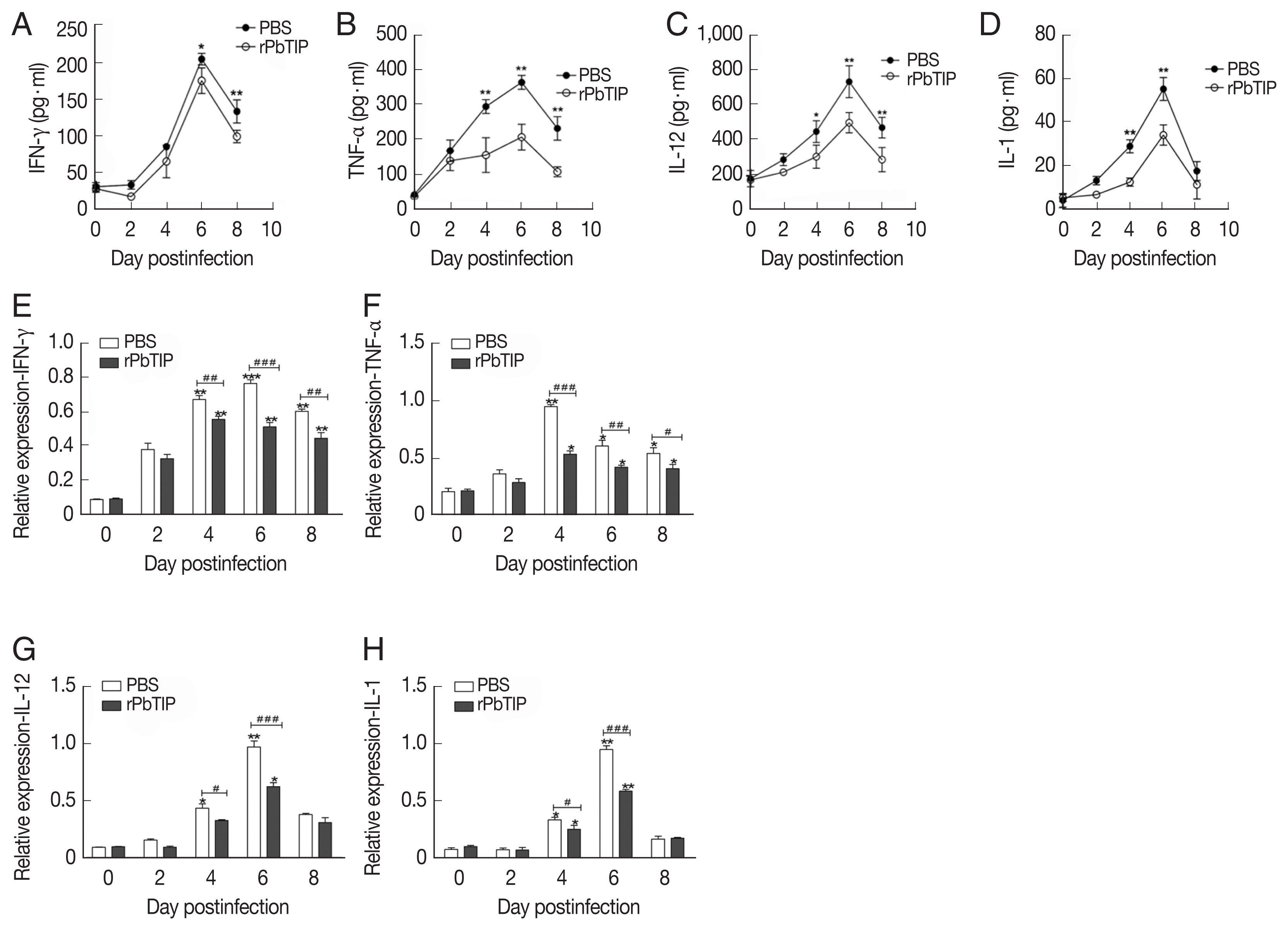
Cytokine levels of IFN-γ, TNF-α, IL-1, IL-12, IL-10, and TGF-β in splenocyte culture or serum were measured by ELISA (R&D Systems, Minneapolis, Minnesota, USA). For quantification of splenocyte cytokines, 106 splenocytes were obtained from 5 mice of each group were isolated on days 0, 2, 4, 6, and 8 p.i. and then cultured for 48 hr ex vivo. Then the culture supernatants or serum were collected and measured by ELISA according to the manufacturer’s protocol. All the experiments were repeated 3 times.
RNA extraction and quantitative real-time-PCR (qRT-PCR)
Five mice from each group were euthanized on days 0, 2, 4, 6, and 8 p.i. Total RNA was extracted from the brain and spleen samples by using TRIzol (Invitrogen). Contaminated DNA was digested by DNAase. Total RNA was transcribed with oligo (dT) primers for reverse transcription (RT) (Takara, Kusatsu, Japan). The resulting cDNA of IFN-γ, TNF-α, IL-1, and IL-12 from the spleen samples and VCAM-1, ICAM-1, CD36, CXCL9, CXCL10, and CXCR3 from the brain samples were used for qRT-PCR assays with SYBR Green quantitative PCR kit (Takara) by 40 cycles in ABI Quant Studio 6 Flex (Applied Biosystems, Foster City, California, USA). Primer sequences were listed in
Table 1. β-actin was used as an internal control for measuring the relative ratio of each target gene. Normal group values were taken as 100% and treated sample values were calculated based on the control. The specificity of qRT-PCR was confirmed by melting-curve analysis. qRT-PCR analysis was determined with 3 replicates.
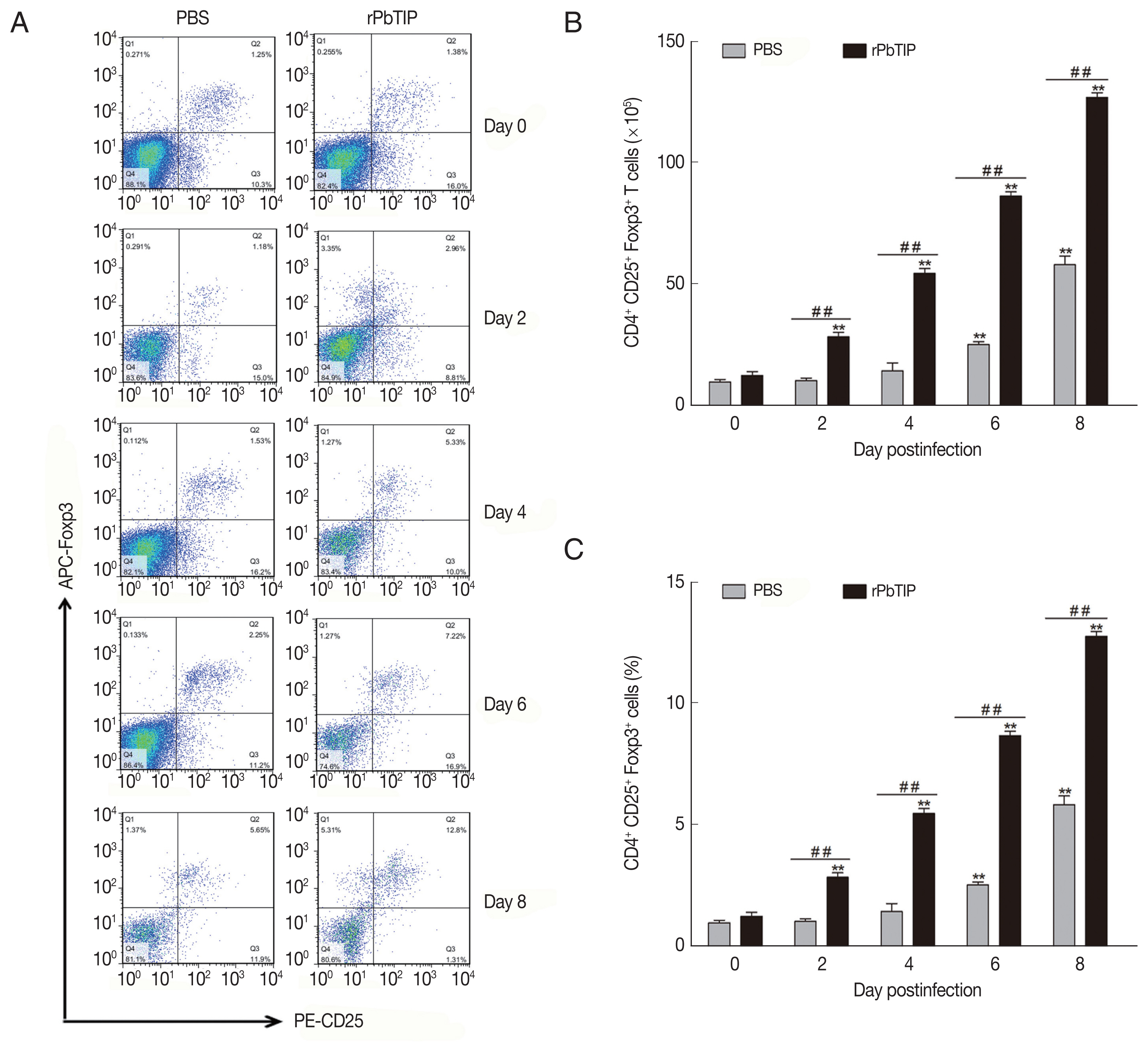
Spleens from 5 mice of each experimental group were harvested on days 0, 2, 4, 6, and 8 p.i. The spleen of day 0 was labeled as control. To measure the activation of regulatory T cells (Tregs), 107 fresh splenocytes were stained with FITC-anti-CD4, and PE-anti-CD25 for surface staining in cell staining buffer (Biolegend, SanDiego, California, USA). After the cells were fixed and permeabilized, intracytoplasmic staining by labeling with fluorochrome conjugated antibody of APC-anti-Foxp3 was performed. After that, the resuspended, fixed and intracellularly labeled cells were analyzed on FACSCalibur flow cytometers (BD Bioscience, San Jose, California, USA). Data were analyzed with FlowJo software (Treestar). The experiments were replicated 3 times.
Statistical methods
For comparisons between 2 groups, statistically significant differences were analyzed by Student’s t-test or 2-way analysis of variance (ANOVA) (SPSS 17.0 software, SPSS Inc., Chicago, Illinois, USA). For comparisons among 3 groups, statistical significance was determined using Kruskal-Wallis test. Significant differences in survival analysis between the groups were calculated by Kaplan-Meier long rank test. Data are presented as means±SEM. P-values<0.05 were considered to be significant.
RESULTS
PbTIP is conserved among Plasmodium species
Through bioinformatic searches from
Plasmodium database in PlasmoDB (
http://www.plasmodb.org), The predicted protein contains a putative signal peptide and transmembrane domain (
Fig. 1A). The PbTIP gene is located on chromosome 12 and encodes a protein of 703 AA with a calculated molecular weight of 80.4 kDa. We identified the
P. berghei ANKA homolog of TIP (PBANKA_124360), which has a similar genome and structure with TIP of
Plasmodium falciparum (
Fig. 1B). Multiple sequence alignments using Clustal W showed that the gene was conserved among
Plasmodium species (
Fig.1B). Based on the above discussion, we annotated this conserved
P. berghei protein as PbTIP.
Anti-rPbTIP antisera determined native PbTIP to be a membrane protein in
P. berghei. We screened PbTIP protein in TMHMM online server (
http://www.cbs.dtu.dk/services/TMHMM/) (
Supplementary Fig. S1A). According to the following criteria, a segment of PbTIP protein corresponding to 204–277 AA were selected for VCBS using Pfam database (
http://pfam.xfam.org/) (
Supplementary Fig. S1B). PbTIP belongs to protein families PF13517 (Family: VCBS). The VCBS of PbTIP protein sequences were also mined in NCBI database (
Supplementary Fig. S1C). Besides that, we also showed SCOP fields among the comparison species. Among such genes and bio-informatics comparisons, we finally selected 204–335 AA for the specific segment for PbTIP. Subsequently, a segment of rPbTIP that corresponds to 204–335 AA was selected to express in
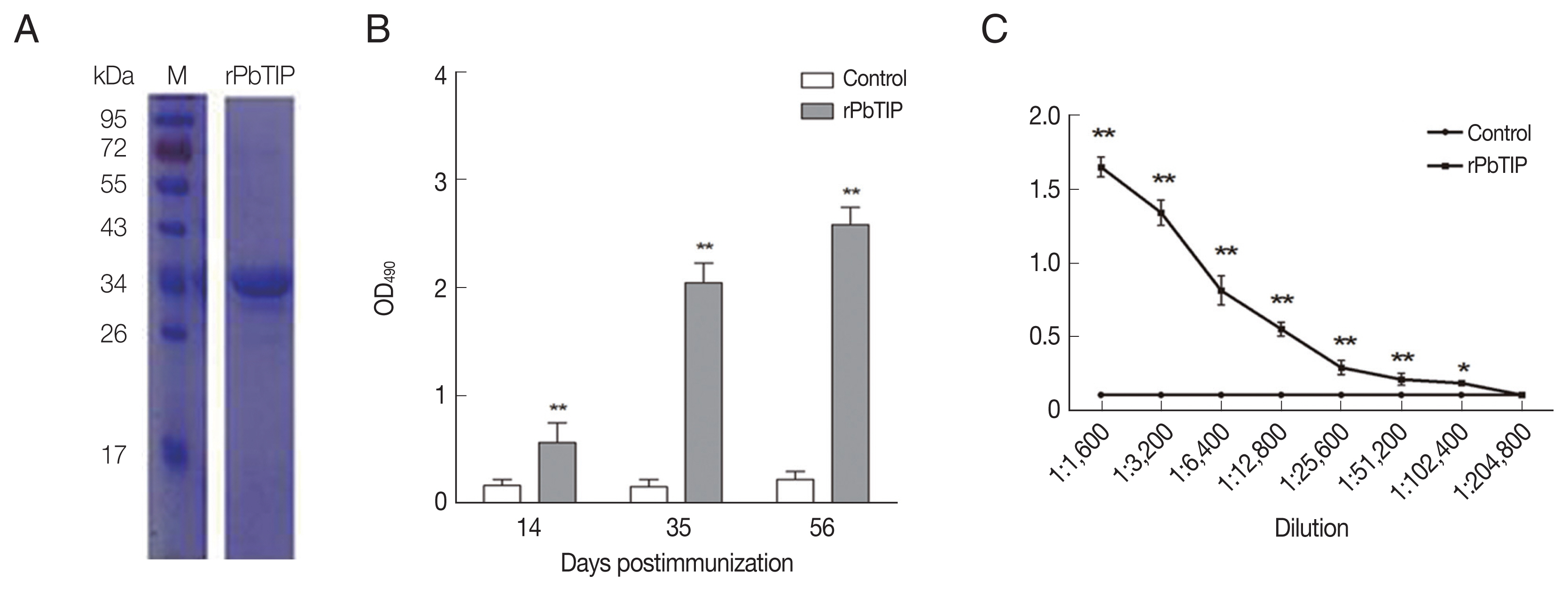
E. coli BL-21. The rPbTIP had a molecular weight of −35 kDa through SDS-PAGE analysis, which was correlated with the predicted molecular size (
Fig. 2A). After ultrafiltration and purification, rPbTIP was used to immunize BALB/c mice to obtain polyclonal antibody. ELISA showed that the mice induced strong antibody responses compared to PBS control (
Fig. 2B) (
t-test,
P<0.01). rPbTIP-specific IgG titers reached 1:204,800 after third immunization (
Fig. 2C) (
t-test,
P<0.01).
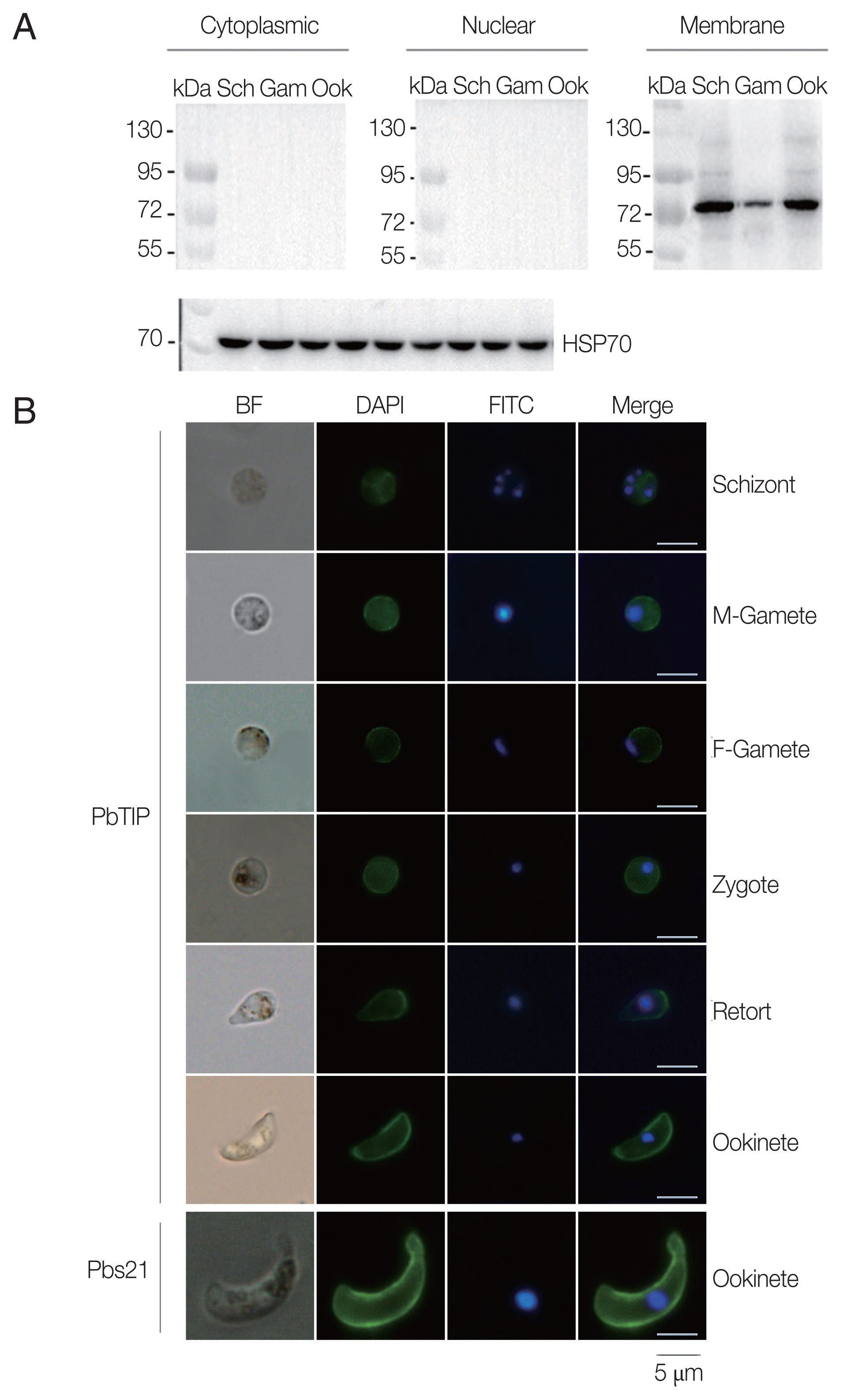
To determine whether the polyclonal antisera can react with native PbTIP in
P. berghei, Western blot was performed with isolated cytoplasmic, nuclear and membrane protein of PbTIP from separated schizonts, gametocytes and ookinetes (
Supplementary Fig. S2). The results demonstrated that the anti-rPbTIP antisera recognized native PbTIP protein bands at approximately 80 kDa as a membrane protein on the outer surface of
P. berghei in 3 representative stages, which was as expected from the PbTIP expression profile (
Fig. 3A). The protein bands confirmed the increased antisera against native PbTIP in
P. berghei, which was consistent with the predicted molecular weight of PbTIP present on the surface of
P. berghei.
Next, we confirmed the expression of PbTIP, and determined the localization of PbTIP by IFA using immune sera of rPbTIP. The consequences revealed strong fluorescence on schizonts, female gametes, male gametes, zygotes, retorts and ookinetes (
Fig. 3B). Pbs21mAb served as a positive control. The localization by IFA indicated that PbTIP was membrane protein on the outer surfaces in asexual and sexual stages of
P. berghei.
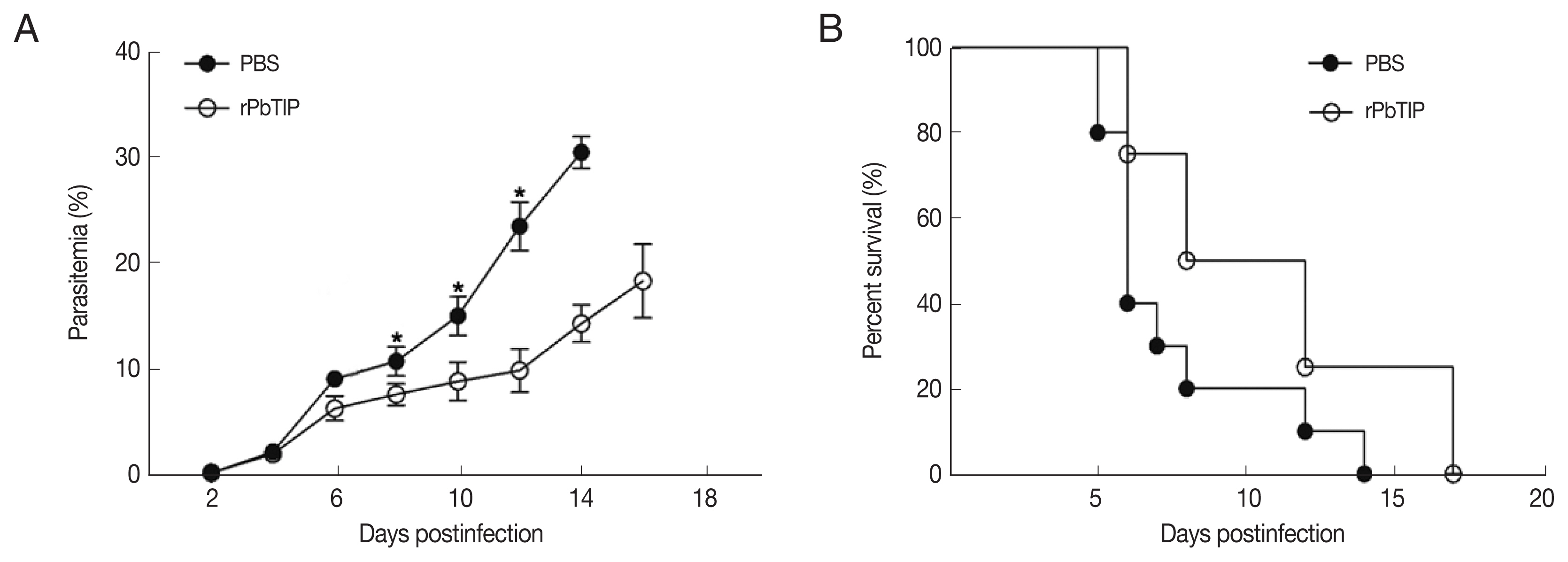
Comparisons of parasitaemia on day 8, 10, and 12 p.i. in the 2 groups demonstrated that parasitaemia in rPbTIP i.v. injection group showed significant difference than parasitaemia in PBS i.v. injection group (2-way ANOVA;
P<0.05) (
Fig. 4A). Nevertheless, comparison of parasitaemia on day 6 p.i. in the 2 groups demonstrated that parasitaemia (6%) in PBS i.v. injection group showed no significant difference than parasitaemia (5.2%) in rPbTIP i.v. injection group (2-way ANOVA;
P>0.05) (
Fig. 4A). These results did not provide any information about the different mortality rates associated with parasitaemia in the 2 groups. Two groups of 20 WT
P. berghei ANKA infected mice each were injected with either PBS or rPbTIP 2 mg/kg on days −1, 0, 1, 3, 5, and 7. We observed rPbTIP i.v. injection group could alleviate the severity of ECM. Two groups demonstrated significant differences in the survival curves by daily monitoring (log-rank test;
P<0.05) (
Fig. 4B). Approximately on day 6, 80% of mice in PBS i.v. injection group demonstrated apparent neurological symptoms and death occurred between days 6 and 7 for ECM. Nevertheless, 30% of mice in rPbTIP i.v. injection group died due to ECM during days 6 to 7 (log-rank test;
P<0.05) (
Fig. 4B).
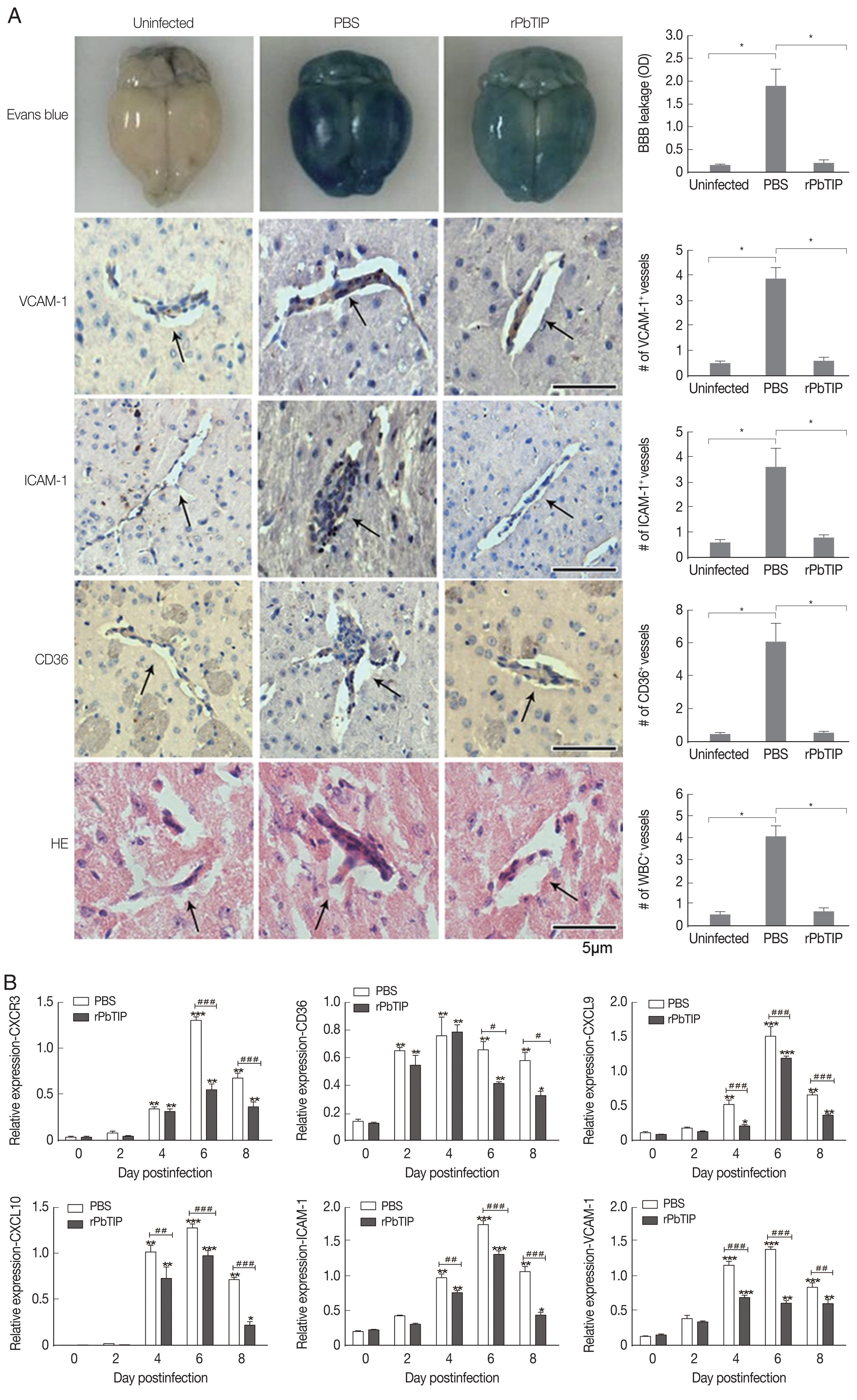
On day 6 of p.i., a substantial loss of Evans blue dye as indicated by BBB leakage in rPbTIP i.v. injection group was lower than that in PBS i.v. injection group (Kruskal-Wallis test;
P<0.05) (
Fig. 5A). Immunostaining of VCAM-1, ICAM-1 and CD36 on the endothelium of microvessels showed less intense in rPbTIP i.v. injection group (Kruskal-Wallis test;
P<0.05) (
Fig. 5A). Immunocytochemistry revealed that the amount of leukocytes per microscopic field in rPbTIP i.v. injection group showed significant reduction than PBS i.v. injection group (Kruskal-Wallis test;
P<0.05) (
Fig. 5A). Then the corresponding adhesive molecules and chemokines were identified by qRT-PCR analysis in the brains on day 6 p.i. qRT-PCR studies have revealed significantly lower expression levels of VCAM-1, ICAM-1 and CD36 in rPbTIP i.v. injection group, which were correlated with the above immunostaining results (2-way ANOVA;
P<0.01). mRNA expression levels of chemokines CXCL9, CXCL10 and receptor CXCR3 were much lower in rPbTIP i.v. injection group (2-way ANOVA;
P<0.01) (
Fig. 5B). These results suggested that rPbTIP i.v. injection group showed significant reduction of inflammation and less severe cerebral pathology than PBS i.v. injection group.
Detection of cytokines in splenocyte culture supernatants by ELISA showed lower splenic cytokine levels in rPbTIP i.v. injection group than PBS i.v. injection group (2-way ANOVA;
P<0.05) (
Fig. 6A–D). Correspondingly, qRT-PCR analysis revealed that the pro-inflammatory cytokines in spleens demonstrated similar results as in ELISA. The results showed smaller number of cytokines in rPbTIP i.v. injection group than that in PBS i.v. injection group (2-way ANOVA;
P<0.05) (
Fig. 6E–H).
Compared with PBS i.v. injection group, CD4
+CD25
+Foxp3
+ regulatory T cells (Tregs) was promoted in rPbTIP i.v. injection group as absolute numbers and percentages (2-way ANOVA;
P<0.01) (
Fig. 7). A few potential mediators of Tregs includes Foxp3, TCR4, CD25, cytotoxic T lymphocyte Ag-4 (CTLA-4), anti-inflammatory cytokines IL-10, and transforming growth factor β (TGF-β) [
24–
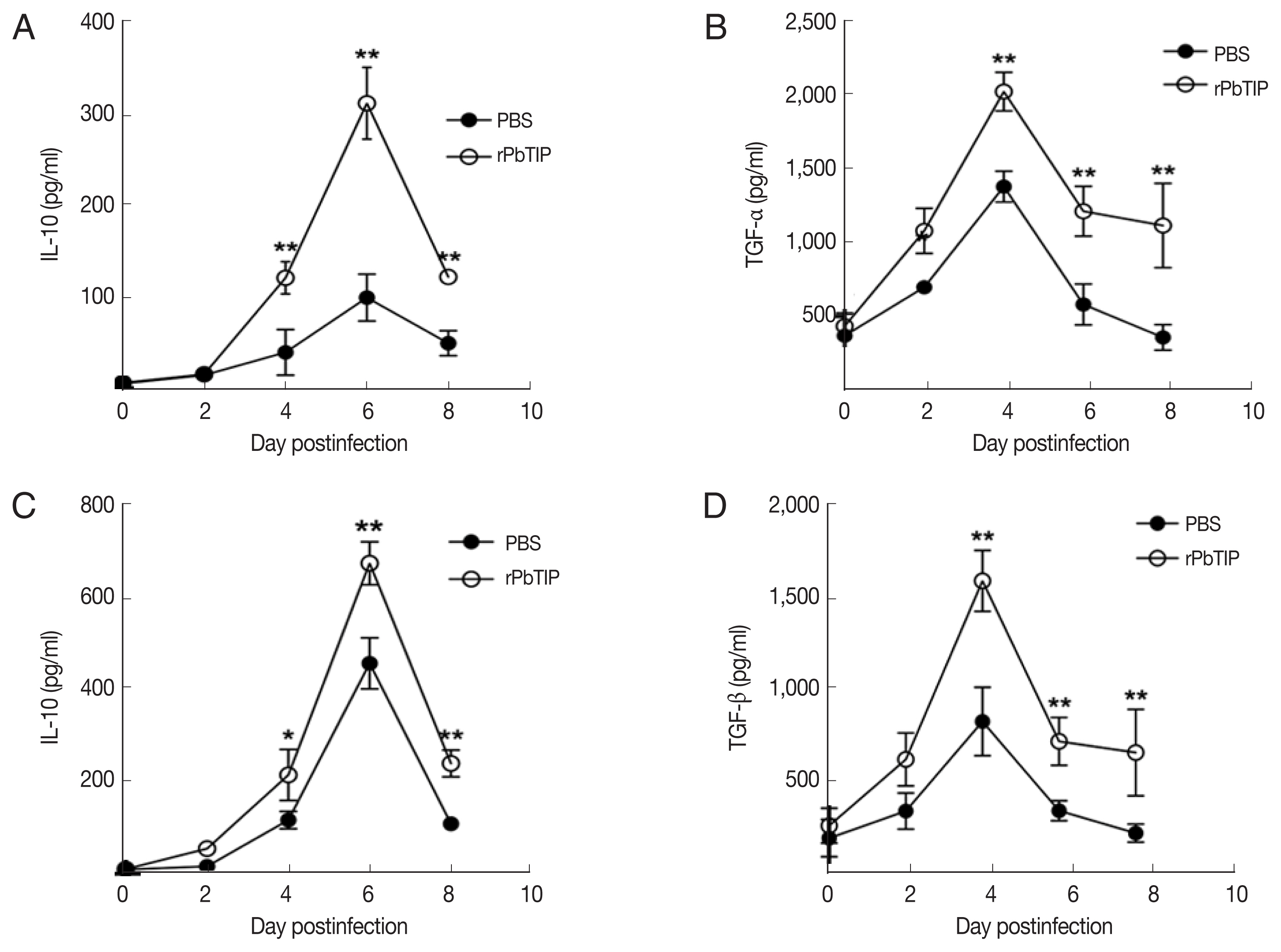
31]. In recent studies, IL-10 and TGF-β have shown essential impact in the regulation of Th1 response. So, we measured the expression of IL-10 and TGF-β in splenocyte culture and serum by ELISA. The results showed that the expression levels of IL-10 and TGF-β from splenocyte culture were elevated in rPbTIP i.v. injection group (2-way ANOVA;
P<0.05) (
Fig. 8A and B). Furthermore, the concentration of IL-10 and TGF-β in sera of rPbTIP i.v. injection group was higher than PBS i.v. injection group (2-way ANOVA;
P<0.05) (
Fig. 8C and D). Collectively, these results suggested that the host immune response from pro-inflammatory milieu to anti-inflammatory milieu was increased by the amount of anti-inflammatory cytokines of Tregs, IL-10 and TGF-β in ECM, reducing the severity of ECM.
DISCUSSION
The mechanism of CM has been a subject of considerable discussion. Recent data from clinical studies suggested that CM has been associated with pathology [
32–
34]. The immune defense to parasite infection is performed by early, intense, proinflammatory cytokine-mediated, immunomodulatory mechanism that eliminate or kill iRBCs. Despite pro-inflammatory was trigged by parasitaemia, there was still a paucity of elimination for parasite, and when excessive can lead to pathology and fatalities. The divergent observations in studies point to the corresponding regulatory response of anti-inflammatory in protective immunity and immune pathogenesis. Therefore, the balance between pro-inflammatory and anti-inflammatory cytokines, the timing, source of anti-inflammatory production, which is induction in determining the final outcome of CM. Overexpression of proinflammtory and intercellular cell adhesion molecule, such as IFN-γ, TNF-α, ICAM-1, and VCAM-1, lead to sequestration of iRBCs and leukocyte to microvascular congestion, resulting in breakdown of the BBB.
Inflammatory and cytoadherence have long been implicated as the pathogenesis with CM in some studies. Therefore, many researchers focus on this aspect. Fiscella et al. [
11] identified a novel factor, T cell immunomodulatory protein (TIP), which can mediate the expression of proinflammatory cytokines to play a protective role in mouse graft-versus-host disease (GVHD) model. In this study we characterized PbTIP, which was highly conserved in
Plasmodium. PbTIP belongs to the protein families PF13517. The localization of PbTIP by IFA was a membrane protein of
Plasmodium and expressed in schizonts, gemetes and ookinetes. We further examined that rPbTIP i.v injection significantly alleviated ECM pathology in C57BL/6 mice and decreased mortality compared with PBS i.v. injection group. Accumulation of rPbTIP may play a role in attenuating immunopathology in ECM. Our study confirmed that excess rPbTIP by i.v injection in ECM was capable of influencing the ongoing process of cerebral pathology.
Several post-mortem examinations have demonstrated that accumulated pRBCs, platelets and leukocytes within the small vessels of brain die due to CM [
35,
36]. The explanation for BBB disruption involves cytoadherence of pRBCs, platelets and leukocytes that obstruct brain microvasculature by increased adhesion molecules [
37,
38]. rPbTIP i.v. injection group showed less BBB leakage than PBS i.v. injection group. In the brain, ICAM-1 acts as a host receptor and is widely distributed on cerebral vessels, which is upregulated by TNF-α and co-localized with pRBCs that die of CM [
39]. CD36, another key endothelial receptor, is expressed on the platelets mediate the clumping of pRBCs [
40]. Our results strongly suggested that the lower levels of these adhesion molecules were expressed in rPbTIP i.v. injection group than PBS i.v. injection group. Accordingly, our results were supported by the detailed histological and immunohistochemistry studies and illustrated that dysregulation of these adhesion molecules did not contribute to the pathogenesis of CM in rPbTIP i.v. injection group.
Cytokines stimulation usually lead to inflammatory infiltration, accumulated endothelial microparticles and correlated with plasma proinflammatory concentration in severe malaria. Severe malaria has been markedly elevated by high circulating levels of IL-1, a pyrogenic cytokine, that induces production of prostaglandins as a key regulator of fever induction [
41]. Meanwhile, it has also been recognized that IL-12 can affect the expression of IFN-γ and TNF-α. During the early stage, the production of IFN-γ may be inclined towards the appropriate elevated levels of IL-12 or IL-18 from dendritic cells or macrophages [
42–
44], which remains a crucial component of proinflammatory responses. Simultaneously, IFN-γ response can up-regulate TNF-α, demonstrating it as a predominant mediator of malaria pathology [
32]. So, we knew that among proinflammatory cytokines, IFN-γ, TNF-α, IL-1, and IL-12 have been implicated in the pathogenesis of CM [
45]. Our data showed that the elicited concentration of proinflammatory cytokines were significantly attenuated in rPbTIP i.v. injection group. So, the peak expression of proinflammatory cytokines were not detected in rPbTIP i.v. injection group compared to PBS i.v. injection group on day 6 of p.i.
The consistent observation regarding timing and balance was related between proinflammatory and anti-inflammatory cytokines. Both of them are important in disease and parasite clearance. Anti-inflammatory cytokines, such as IL-10, and TGF-β induced a protective or counter-regulatory effect [
46]. There is already existing evidence that marine malaria was associated with high plasma levels of TNF-α [
47,
48] and low concentrations of TGF-β [
49,
50] during the course of increased disease severity. Experiments were ongoing to determine the anti-inflammatory cytokines (IL-10 and TGF-β) in our study. There was an extremely significant finding that high ratio of circulating IL-10 and TGF-β levels were consistently found in rPbTIP i.v. injection group compared to PBS i.v. injection group.
The potential mediators of Tregs have been demonstrated to be Foxp3, TCR4, CD25, cytotoxic T lymphocyte Ag-4 (CTLA-4) and anti-inflammatory cytokines IL-10, TGF-β [
24–
31]. Tregs are only present in CD4
+CD25
+Foxp3
+Treg cells. The Foxp3 gene can transfer the naive T cells into Tregs [
29,
30]. So, we prospectively studied the relationship between Tregs and anti-inflammatory cytokines. It was equally surprising that Tregs were much more apparent in WT
P. berghei ANKA infected mice by i.v. injection of rPbTIP. In support of this, we have showed that the increase in anti-inflammatory ratio was associated with an increase in Tregs, which may modify the cerebral environments from overwhelming proinflammatory to anti-inflammatory. Thus, we postulated that Tregs and anti-inflammatory cytokines (IL-10 and TGF-β) act as indicators and were resistant to ECM due to decreased proinflammatory cascade, leading to severe pathology within the brain [
51].
In summary, our studies sought to assess the function and influence of PbTIP on the outcome of ECM. Interestingly, the presence of TIP homologues in P. berghei has generated interest in assessing its contribution to uncontrolled pro-inflammatory cytokines that are associated with excess immunopathology of ECM. Results demonstrated that increased expression of recombinant PbTIP for blood-stage malaria parasites were able to resist ECM, as evidenced by reduction in cerebral inflammation and parasitaemia. The results on the outcome of P. berghei malaria in PbTIP-immunized mice were probably unanticipated. Additional studies of Plasmodium TIP were required to further assess its role on severe complications of ECM. In the midst of stimulation of all new studies, it was observed that no new approach has been proven to be effective in reducing the mortality of ECM. PbTIP may act as a novel example for difficulty in separating immunopathology and its effect in the ECM studies.
Notes
-
CONFLICT OF INTEREST
All authors disclose that there was no conflict of interest of any type from the beginning of the work to the submission of manuscript.
Supplementary Information
Supplementary Fig. S1.
Identification of expressed segments of PbTIP. (A) PbTIP protein was screened in TMHMM online sever. (B) The results showed VCBS field from 204–277 AA using SMART online server. (C) VCBS field are mined in NCBI database, corresponding to 204–335 AA.
Supplementary Fig. S2.
The extraction and purification of schizonts, gametocytes and ookinetes from P. berghei ANKA.
ACKNOWLEDGMENT
This work was supported by grant from the National Natural Science Foundation of China (81471978).
Fig. 1Bioinformatic analyses of PbTIP. (A) Schematic diagram of PbTIP showing the total length of PbTIP, a signal peptide and transmembrane domain. The E. coli expression segment shows 204–335 AA. (B) Alignment of protein sequences of PbTIP in Plasmodium species: P. berghei (Pb), P. vivax (Pv), P. falciparum (Pf), and P. yoelii (Py).

Fig. 2Recombinant PbTIP protein purification and immunization. (A) rPbTIP was purified and separated on 10% SDS-PAGE gel. (B) rPbTIP-antibody titers in immunized BALB/c mice. Serum samples from immunized and control mice were collected post-immunization on days 14, 35, and 56. The data represented 3 separate experiments. Error bars were presented as mean±SD. SD indicates the assay variance. (C) ELISA showing total IgG titer of anti-rPbTIP at 10 days. Mean of control antisera±3×SD represents the broken lines. IgG titers correspond to the last dilution of anti-rPbTIP sera in which the OD490 values were above the cut-off value. The cut-off value was defined as the pooled sera from control mice. Results are representative of 3 independent experiments. M, molecular weight marker. **P<0.01 (Student’s t-test).
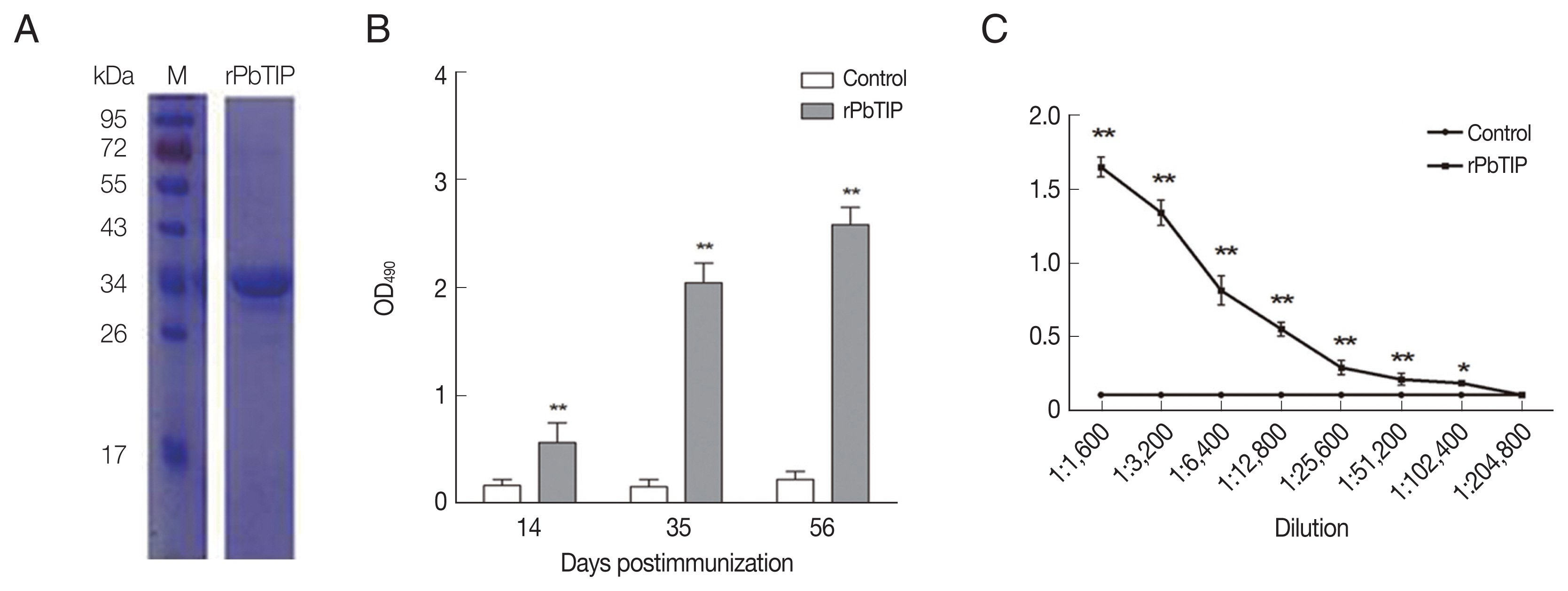
Fig. 3Expression and localization of PbTIP in the parasites. (A) Western blot of PbTIP in asexual and sexual stage parasites. Parasite antigens from isolated cytoplasmic, nuclear and membrane protein (30 μg/lane) of schizonts, gametocytes and ookinetes were incubated with anti-rPbTIP antisera (1:100). Anti-HSP70 was used as positive control. (B) Blood smears from parasite samples were stained at different stages after collecting the P. berghei infected blood. Parasites were fixed and stained with anti-rPbTIP sera and then with FITC-conjugated goat anti-mouse IgG (green). Nuclei were labeled with DAPI (blue). Positive control-Pbs21 mAb. Scale-bars: 5 μm. BF, bright field.
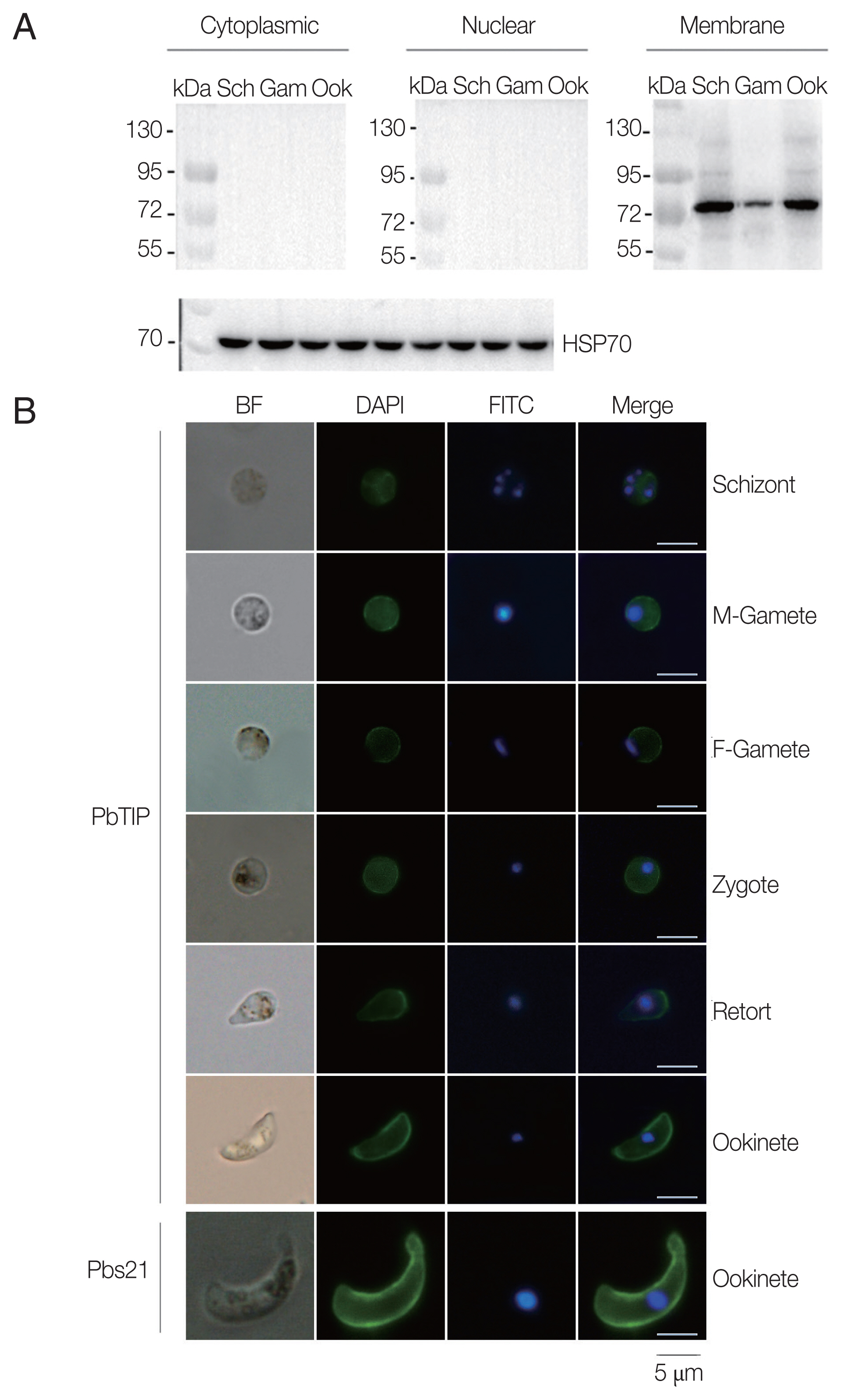
Fig. 4rPbTIP i.v. injection group showed improved host survival. (A) Dynamics of parasitaemia was monitored for 17 days. (B) Cumulative survival analysis. rPbTIP i.v. injection group and PBS i.v. injection group were monitored daily for survival. *P<0.05; log-rank test.
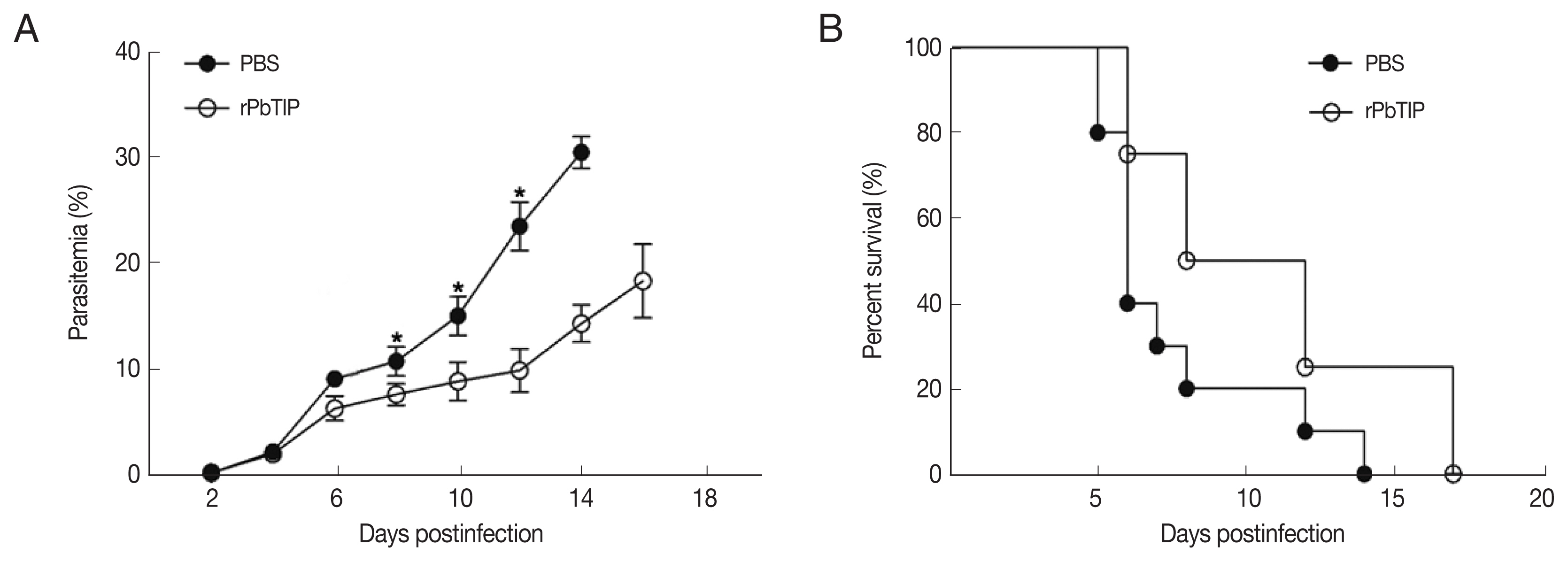
Fig. 5(A) i.v. injection of rPbTIP reduced endothelium activation and BBB leakage. On day 6 p.i., infected mice from each group were processed for histology using HE staining and immunohistochemical analysis with anti-VCAM-1, ICAM-1 and CD36 antibodies. The Kruskal-Wallis test was used for 3-group comparisons, and Dunn’s test was employed for post hoc analysis. Top panel: Representative brain images showing the extent of vascular leakage, while the corresponding bar graphs showing the quantification of BBB leakage. Significant difference was observed between PBS i.v. injection group and rPbTIP i.v. injection group (*P<0.05). Bottom 4 panels: Representative images of brain sections with the microvessels (arrows), and accordingly the bar graphs indicate quantification of VCAM-1, ICAM-1, CD36, and leukocytes-positive microvessels. Microvessels per microscopic field were quantified in 20 fields per mouse, values are presented as mean±SEM from 5 mice of each group. (B) Quantitation of mRNA levels of adhesive molecules and chemokines in brain tissues by qRT-PCR. On day 0 and days 2, 4, 6, and 8 post-infection, 5 mice were used for each group. Results are representatives of 3 independent experiments. Total RNA was extracted from the brains, qRT-PCR with β-actin as the internal reference. *P<0.05, and **P<0.01 indicate the comparison with the uninfected group. #P<0.05 and ##P<0.01 for comparison between rPbTIP i.v. injection group and PBS group (2-way ANOVA).
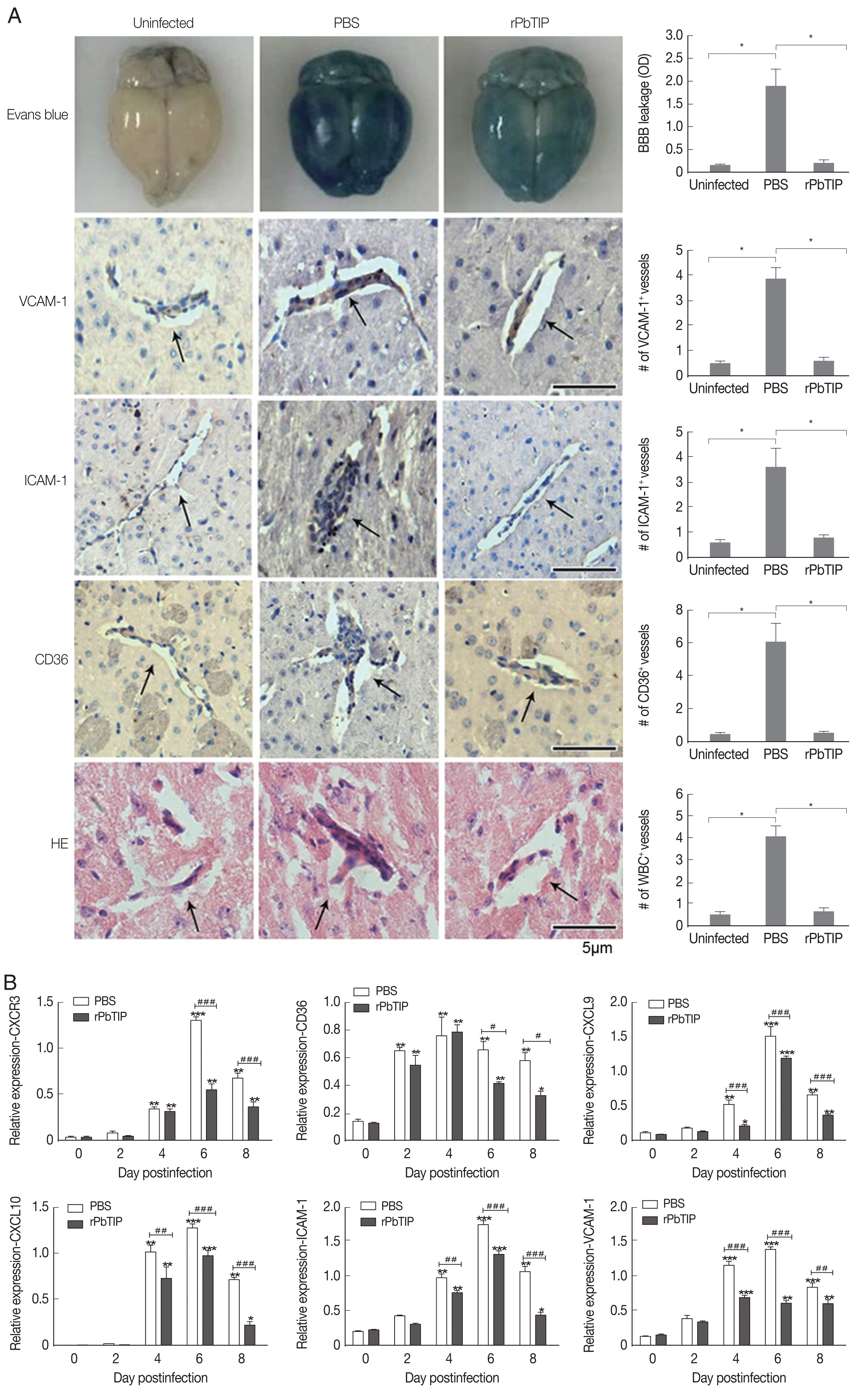
Fig. 6Proteins and transcripts of proinflammatory cytokines in spleens. (A–D) The levels of IFN-γ, TNF-α, IL-1, and IL-12 in splenocyte culture supernatants were measured by ELISA. Each experiment was repeated 3 times. Values are presented as mean±SEM for 5 mice in each group. *P<0.05, and **P<0.01, for comparison between rPbTIP i.v. injection group and PBS mice. (E–H) Quantitation of mRNA levels of pro-inflammatory cytokines in spleens by qRT-PCR. On day 0 and days 2, 4, 6, and 8 post-infection, 5 mice were used for each group. Results are representatives of 3 independent experiments. Total RNA was extracted from the spleens, and relative expression of individual genes was quantified with β-actin as the internal reference. *P<0.05, and **P<0.01 indicate the comparison with the uninfected group. #P<0.05 and ##P<0.01 for comparison between WT P. berghei ANKA infected by i.v injection of rPbTIP and PBS groups (2-way ANOVA).
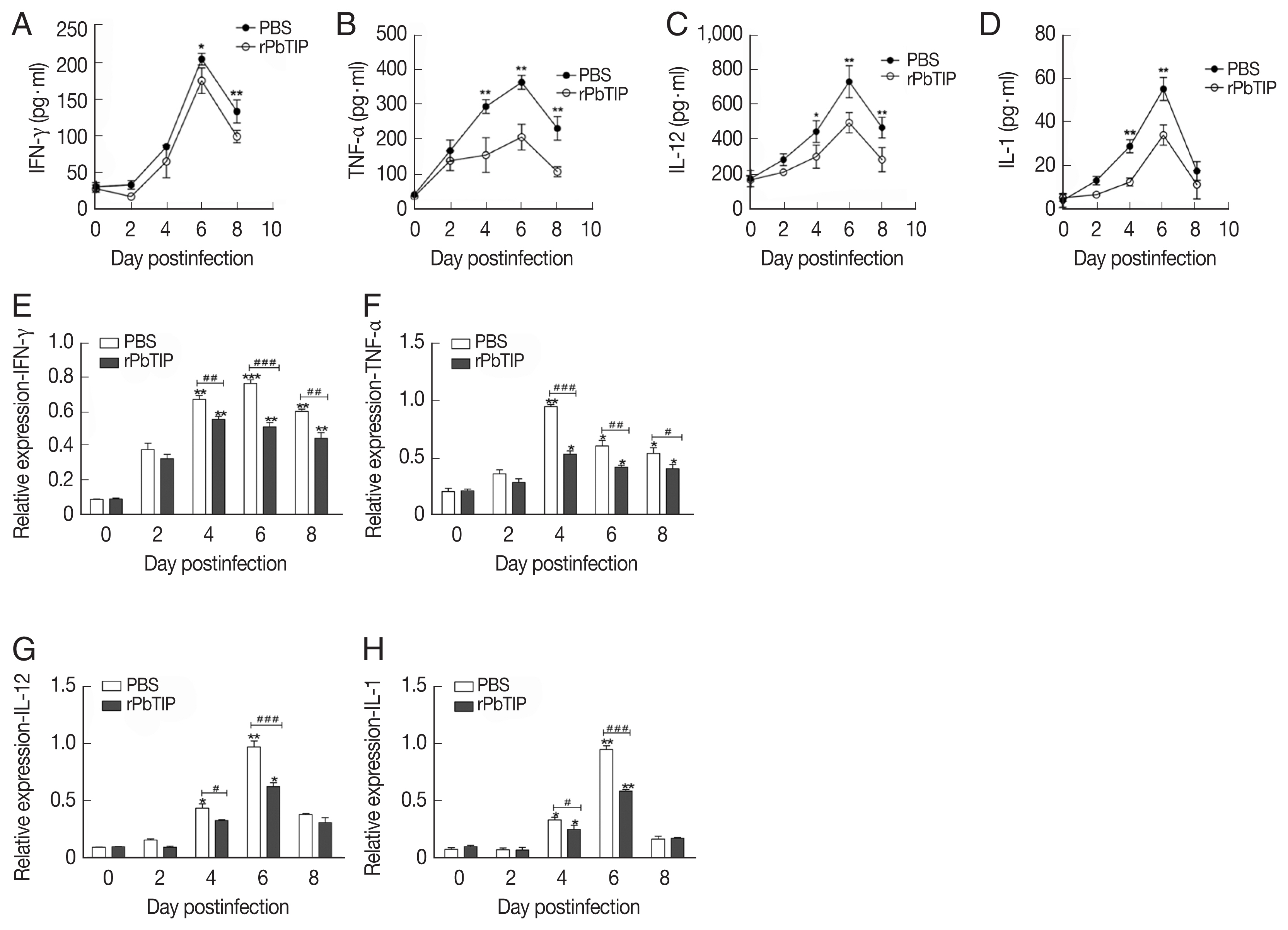
Fig. 7The expansion of Tregs and increased production of anti-inflammatory cytokines (IL-10 and TGF-β). (A–C) Tregs were quantified by flow cytometry. rPbTIP i.v. elicited the expansion of Tregs. (A) Representative dot plots of Tregs in spleens was detected by flow cytometry. (B, C) The absolute numbers and the proportion of Tregs. On day 0 and days 2, 4, 6, and 8 post-infection, 5 mice were euthanized for each group. Results are representatives of 3 independent experiments. Value sare presented as mean±SEM. *P<0.05, and **P<0.01 indicate the comparison with the uninfected group. #P<0.05 and ##P<0.01 for comparison between rPbTIP i.v. injection group and PBS group (2-way ANOVA).
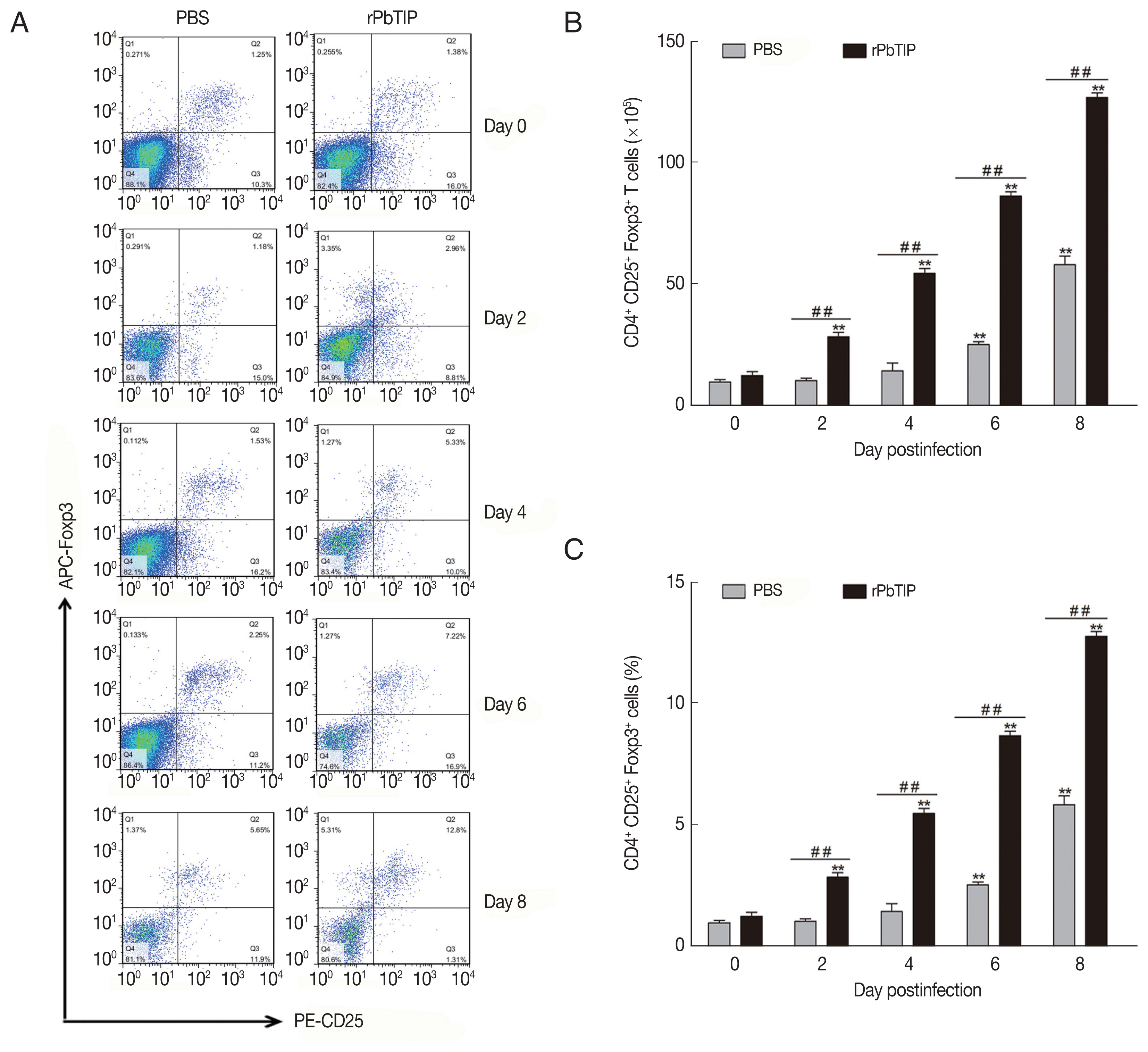
Fig. 8The concentrations of IL-10 and TGF-β in serum cultured splenocytes and serum by ELISA. Both cytokine levels were harvested from the same culture supernatant and serum. (A, B) IL-10 and TGF-β were determined in cultured splenocytes. (C, D) IL-10 and TGF-β were measured in the serum. Results are representatives of 3 independent experiments. Data were presented as mean±SEM. *P<0.05, and **P<0.01 for comparison between WT P. berghei ANKA infected by i.v injection of rPbTIP and PBS groups (2-way ANOVA).
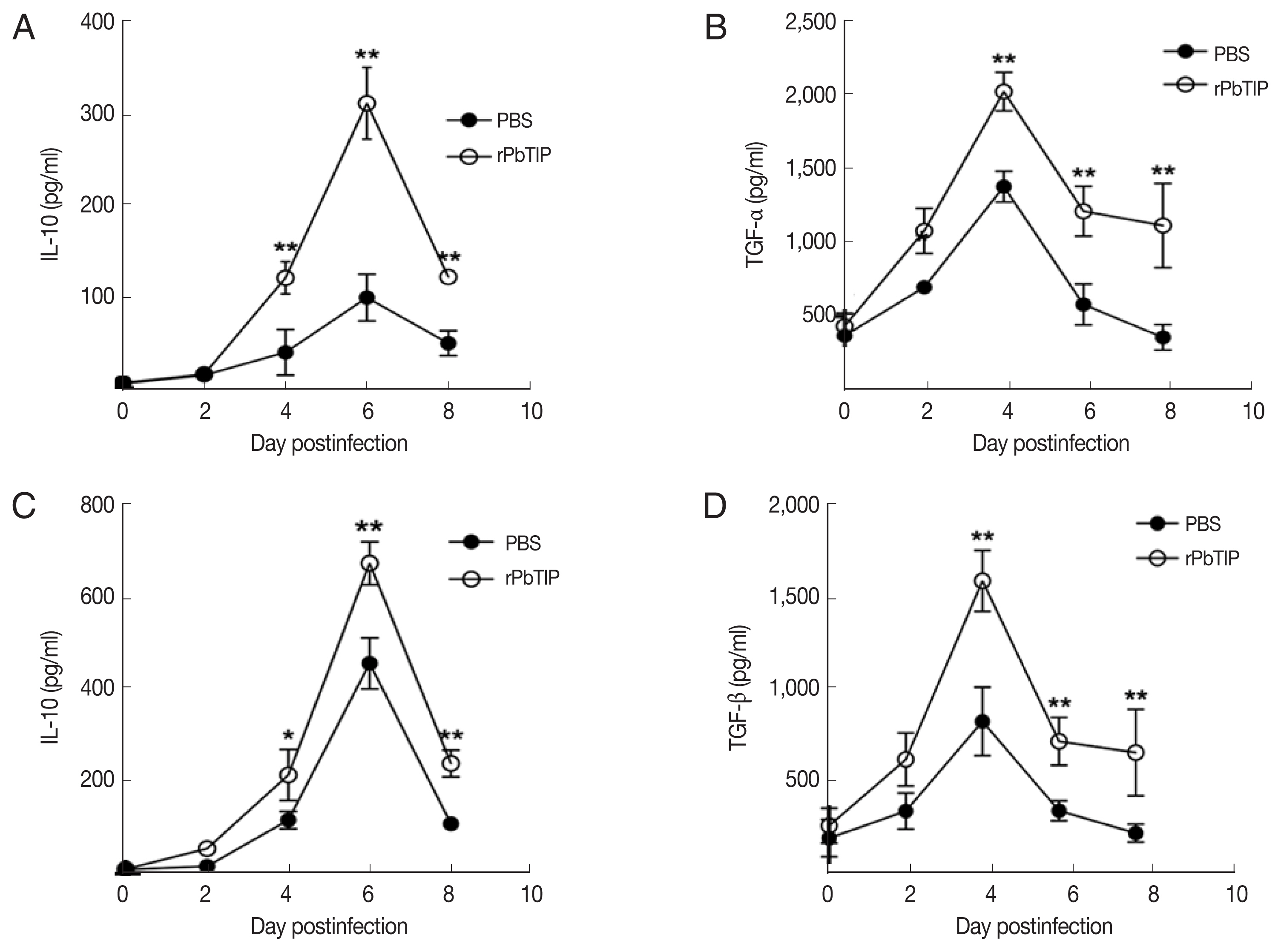
Table 1Primers for quantitative real-time PCR
Table 1
|
Target gene |
Forward |
Reverse |
|
β-actin |
GATTACTGCTCTGGCTCCTAGC |
GACTCATCGTACTCCTGCTTGC |
|
ICAM-1 |
GGCAGCAAGTAGGCAAGGAC |
CTGGCGGCTCAGTATCTCCT |
|
VCAM-1 |
CCTCACTTGCAGCACTACGG |
CATGGTCAGAACGGACTTGG |
|
CD36 |
CCCTCCAGAATCCAGACAAC |
CACAGGCTTTCCTTCTTTGC |
|
CXCL9 |
CCGAGGCACGATCCACTACA |
AGTCCGGATCTAGGCAGGTTTG |
|
CXCL10 |
GCCGTCATTTTCTGCCTCAT |
GCTTCCCTATGGCCCTCATT |
|
CXCR3 |
AAGTGCCAAAGGCAGAGAAG |
AAAGTCCGAGGCATCTAGCA |
|
IL-12 |
TTTGCTGGTGTCTCCACTCA |
CATCTTCTTCAGGCGTGTCA |
|
IL-1β |
GCTGCTTCCAAACCTTTGAC |
AGCTTCTCCACAGCCACAAT |
|
TNF-α |
TATGGCTCAGGGTCCAACTC |
CCCATTTGAGTCCTTGATGG |
|
IFN-γ |
TGATCCTTTGGACCCTCTGA |
ACAGCCATGAGGAAGAGCTG |
|
TGF-β |
AGCAACAATTCCTGGCGTTACCTT |
CCTGTATTCCGTCTCCTTGGTTCAG |
|
IL-10 |
CTATGCTGCCTGCTCTTACTGACTG |
GAGTCGGTTAGCAGTATGTTGTCCAG |
References
- 1. World Health Organization. World Malaria Report, 2018. Geneva, Switzerland. World Health Organization. 2018.
- 2. White NJ, Pukrittayakamee S, Hien TT, Faiz MA, Mokuolu OA, Dondorp AM. Malaria. Lancet 2014;383:723-735.
- 3. John CC, Bangirana P, Byarugaba J, Opoka RO, Idro R, Jurek AM, Wu B, Boivin MJ. Cerebral malaria in children is associated with long-term cognitive impairment. Pediatrics 2008;122:92-99.
- 4. MacCormick IJC, Beare NA, Taylor TE, Barrera V, White VA, Hiscott P, Molyneux ME, Dhillon B, Harding SP. Cerebral malaria in children: using the retina to study the brain. Brain 2014;137:2119-2142.
- 5. Guo J, Wakninegrinberg JH, Mitchell AJ, Barenholz Y, Golenser J. Reduction of experimental cerebral malaria and its related proinflammatory responses by the novel liposome-based β-methasone nanodrug. Biomed Res Int 2014;2014:66-70.
- 6. Khandare AV, Bobade D, Deval M, Patil T, Saha B, Prakash D. Expression of negative immune regulatory molecules, pro-inflammatory chemokine and cytokines in immunopathology of ECM developing mice. Acta Trop 2017;172:58-63.
- 7. Dunst J, Kamena F, Matuschewski K. Cytokines and Chemokines in Cerebral Malaria Pathogenesis. Front Cell Infect Microbiol 2017;7:324.
- 8. Mathieu C, Demarta-Gatsi C, Porcherie A, Brega S, Thiberge S, Ronce K, Smith L, Peronet R, Amino R, Ménard R, Mécheri S.
Plasmodium berghei histamine-releasing factor favours liver-stage development via inhibition of IL-6 production and associates with a severe outcome of disease. Cell Microbiol 2015;17:542-558.
- 9. Keita Alassane S, Nicolau-Travers ML, Menard S, Andreoletti O, Cambus JP, Gaudre N, Wlodarczyk M, Blanchard N, Berry A, Abbes S, Colongo D, Faye B, Augereau JM, Lacroux C, Iriart X, Benoit-Vical F. Young Sprague Dawley rats infected by Plasmodium berghei: A relevant experimental model to study cerebral malaria. PLoS One 2017;12:e0181300.
- 10. Pino P, Vouldoukis I, Kolb JP, Mahmoudi N, Desportes-Livage I, Bricaire F, Danis M, Dugas B, Mazier D.
Plasmodium falciparum--infected erythrocyte adhesion induces caspase activation and apoptosis in human endothelial cells. J Infect Dis 2003;187:1283-1290.
- 11. Fiscella M, Perry JW, Teng B, Bloom M, Zhang C, Leung K, Pukac L, Florence K, Concepcion A, Liu B, Meng Y, Chen C, Elgin EC, Kanakaraj P, Kaufmann TE, Porter J, Cibotti R, Mei Y, Zhou J, Chen G, Roschke V, Komatsoulis G, Mansfield B, Ruben S, Sanyal I, Migone TS. TIP, a T-cell factor identified using high-throughput screening increases survival in a graft-versus-host disease model. Nat Biotechnol 2003;21:302-307.
- 12. Nono JK, Lutz MB, Brehm K. EmTIP, a T-Cell Immunomodulatory Protein Secreted by the Tapeworm Echinococcus multilocularis Is Important for Early Metacestode Development. PLoS Neglect Trop Dis 2014;8:e2632.
- 13. Blagborough AM, Sinden RE.
Plasmodium berghei HAP2 induces strong malaria transmission-blocking immunity in vivo and in vitro. Vaccine 2009;27:5187-5194.
- 14. Chan JA, Fowkes FJ, Beeson JG. Surface antigens of Plasmodium falciparum-infected erythrocytes as immune targets and malaria vaccine candidates. Cell Mol Life Sci 2014;71:3633-3657.
- 15. Dempsey E, Prudêncio M, Fennell BJ, Gomes-Santos CS, Barlow JW, Bell A. Antimitotic herbicides bind to an unidentified site on malarial parasite tubulin and block development of liver-stage Plasmodium parasites. Mol Biochem Parasit 2013;188:116-127.
- 16. Deligianni E, Morgan RN, Bertuccini L, Kooij TW, Laforge A, Nahar C, Poulakakis N, Schüler H, Louis C, Matuschewski K, Siden-Kiamos I. Critical role for a stage-specific actin in male exflagellation of the malaria parasite. Cell Microbiol 2011;13:1714-1730.
- 17. Eisenhaber B, Maurer-Stroh S, Novatchkova M, Schneider G, Eisenhaber F. Enzymes and auxiliary factors for GPI lipid anchor biosynthesis and post-translational transfer to proteins. Bioessays 2003;25:367-385.
- 18. Baccarella A, Huang BW, Fontana MF, Kim CC. Loss of Toll-like receptor 7 alters cytokine production and protects against experimental cerebral malaria. Malaria J 2014;13:354.
- 19. He X, Yan J, Zhu X, Wang Q, Pang W, Qi Z, Wang M, Luo E, Parker DM, Cantorna MT, Cui L, Cao Y. Vitamin D inhibits the occurrence of experimental cerebral malaria in mice by suppressing the host inflammatory response. J Immunol 2014;193:1314-1323.
- 20. Mlambo G, Kumar N, Yoshida S. Functional immunogenicity of baculovirus expressing Pfs25, a human malaria transmission-blocking vaccine candidate antigen. Vaccine 2010;28:7025-7029.
- 21. Kumar R, Ray PC, Datta D, Bansal GP, Angov E, Kumar N. Nanovaccines for malaria using Plasmodium falciparum antigen Pfs25 attached gold nanoparticles. Vaccine 2015;33:5064-5071.
- 22. Wei X, Li Y, Sun X, Zhu X, Feng Y, Liu J, Jiang Y, Shang H, Cui L, Cao Y. Erythropoietin protects against murine cerebral malaria through actions on host cellular immunity. Infect Immun 2014;82:165-173.
- 23. Yamakuchi M, Kirkiles-Smith NC, Ferlito M, Cameron SJ, Bao C, Fox-Talbot K, Wasowska BA, Baldwin WM 3rd, Pober JS, Lowenstein CJ. Antibody to human leukocyte antigen triggers endothelial exocytosis. Proc Natl Acad Sci USA 2007;104:1301-1306.
- 24. Wahl SM, Vázquez N, Chen W. Regulatory T cells and transcription factors: gatekeepers in allergic inflammation. Curr Opin Immunol 2004;16:768-774.
- 25. Sakaguchi S. Naturally arising CD4+ regulatory t cells for immunologic self-tolerance and negative control of immune responses. Annu Rev Immunol 2004;22:531-562.
- 26. Sakaguchi S. Naturally arising Foxp3-expressing CD25+CD4+ regulatory T cells in immunological tolerance to self and non-self. Nat Immunol 2005;6:345-352.
- 27. McHugh RS, Whitters MJ, Piccirillo CA, Young DA, Shevach EM, Collins M, Byrne MC. CD4+CD25+ immunoregulatory T cells: gene expression analysis reveals a functional role for the glucocorticoid-induced TNF receptor. Immunity 2002;16:311-323.
- 28. Chen W, Jin W, Wahl S. Engagement of cytotoxic T lymphocyte–associated antigen 4 (CTLA-4) induces transforming growth factor β (TGF-β) production by murine CD4 T Cells. J Exp Med 1998;188:1849-1857.
- 29. Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol 2003;4:330-336.
- 30. Khattri R, Cox T, Yasayko SA, Ramsdell F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol 2003;4:337-342.
- 31. Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science 2003;299:1057-1061.
- 32. Amani V, Vigário AM, Belnoue E, Marussig M, Fonseca L, Mazier D, Rénia L. Involvement of IFN-gamma receptor-medicated signaling in pathology and anti-malarial immunity induced by Plasmodium berghei infection. Eur J Immunol 2000;30:1646.
- 33. Belnoue E, Potter SM, Rosa DS, Mauduit M, Grüner AC, Kayibanda M, Mitchell AJ, Hunt NH, Rénia L. Control of pathogenic CD8+ T cell migration to the brain by IFN-gamma during experimental cerebral malaria. Parasite Immunol 2010;30:544-553.
- 34. Grau GE, Heremans H, Piguet PF, Pointaire P, Lambert PH, Billiau A, Vassalli P. Monoclonal antibody against interferon gamma can prevent experimental cerebral malaria and its associated overproduction of tumor necrosis factor. Proc Natl Acad Sci USA 1989;86:5572-5574.
- 35. Turner GD, Morrison H, Jones M, Davis TM, Looareesuwan S, Buley ID, Gatter KC, Newbold CI, Pukritayakamee S, Nagachinta B, White NJ, Berendt AR. An immunohistochemical study of the pathology of fatal malaria. Evidence for widespread endothelial activation and a potential role for intercellular adhesion molecule-1 in cerebral sequestration. Am J Pathol 1994;145:1057-1069.
- 36. Roberts DJ, Pain A, Kai O, Kortok M, Marsh K. Autoagglutination of malaria-infected red blood cells and malaria severity. Lancet 2000;355:1427-1428.
- 37. Rénia L, Howland SW, Claser C, Charlotte GA, Suwanarusk R, Hui TT, Russell B, Ng LF. Cerebral malaria: mysteries at the blood-brain barrier. Virulence 2012;3:193-201.
- 38. Medana IM, Turner GD. Human cerebral malaria and the blood-brain barrier. Int J Parasitol 2006;36:555-568.
- 39. Turner GD, Ly VC, Nguyen TH, Tran TH, Nguyen HP, Bethell D, Wyllie S, Louwrier K, Fox SB, Gatter KC, Day NP, Tran TH, White NJ, Berendt AR. Systemic endothelial activation occurs in both mild and severe malaria. Correlating dermal microvascular endothelial cell phenotype and soluble cell adhesion molecules with disease severity. Am J Pathol 1998;152:1477-1487.
- 40. Pain A, Ferguson DJ, Kai O, Urban BC, Lowe B, Marsh K, Roberts DJ. Platelet-mediated clumping of Plasmodium falciparum-infected erythrocytes is a common adhesive phenotype and is associated with severe malaria. Proc Natl Acad Sci USA 2001;98:1805-1810.
- 41. Netea MG, Kullberg BJ, Van der Meer JW. Circulating cytokines as mediators of fever. Clin Infect Dis 2000;31( suppl):178-184.
- 42. Maitland K1, Levin M, English M, Mithwani S, Peshu N, Marsh K, Newton CR. Severe P. falciparum malaria in Kenyan children: evidence for hypovolaemia. QJM 2003;96:427-434.
- 43. Perkins DJ, Kremsner PG, Weinberg JB. Inverse relationship of plasma prostaglandin E2 and blood mononuclear cell cyclooxygenase-2 with disease severity in children with Plasmodium falciparum malaria. J Infect Dis 2001;183:113-118.
- 44. Schofield L, Hackett F. Signal transduction in host cells by a glycosylphosphatidylinositol toxin of malaria parasites. J Exp Med 1993;177:145-153.
- 45. Olaniyan SA, Amodu OK, Bakare AA, Troye-Blomberg Marita, Omotade OO, Rockett KA; MalariaGEN Consortium. Tumour necrosis factor alpha promoter polymorphism, TNF −238 is associated with severe clinical outcome of falciparum malaria in Ibadan southwest Nigeria. Acta Trop 2016;161:62-67.
- 46. Clark IA, Cowden WB. The pathophysiology of falciparum malaria. Pharmacol Ther 2003;99:221-260.
- 47. Grau GE, Taylor TE, Molyneux ME, Wirima JJ, Vassalli P, Hommel M, Lambert PH. Tumor necrosis factor and disease severity in children with falciparum malaria. New Engl J Med 1989;320:1586-1591.
- 48. Kwiatkowski D, Hill AV, Sambou I, Twumasi P, Castracane J, Manogue KR, Cerami A, Brewster DR, Greenwood BM. TNF concentration in fatal cerebral, non-fatal cerebral, and uncomplicated Plasmodium falciparum malaria. Lancet 1990;336:1201-1204.
- 49. Perkins DJ, Weinberg JB, Kremsner PG. Reduced interleukin-12 and transforming growth factor—β1 in severe childhood malaria: relationship of cytokine balance with disease severity. J Infect Dis 2000;182:988-992.
- 50. Wenisch C, Parschalk B, Burgmann H, Looareesuwan S, Graninger W. Decreased serum levels of TGF-beta in patients with acute Plasmodium falciparum malaria. J Clin Immunol 1995;15:69-73.
- 51. Keswani T, Sarkar S, Sengupta A, Bhattacharyya A. Role of TGF-β and IL-6 in dendritic cells, Treg and Th17 mediated immune response during experimental cerebral malaria. Cytokine 2016;88:154-166.