Abstract
Echinostoma mekongi was reported as a new species in 2020 based on specimens collected from humans in Kratie and Takeo Province, Cambodia. In the present study, its metacercarial stage has been discovered in Filopaludina martensi cambodjensis snails purchased from a local market nearby the Tonle Sap Lake, Pursat Province, Cambodia. The metacercariae were fed orally to an experimental hamster, and adult flukes were recovered at day 20 post-infection. They were morphologically examined using light and scanning electron microscopes and molecularly analyzed by sequencing of their mitochondrial cox1 and nad1 genes. A total of 115 metacercariae (1–8 per snail) were detected in 60 (60.0%) out of 100 Filopaludina snails examined. The metacercariae were round, 174 μm in average diameter (163–190 μm in range), having a thin cyst wall, a head collar armed with 37 collar spines, and characteristic excretory granules. The adult flukes were elongated, ventrally curved, 7.3 (6.4–8.2)×1.4 (1.1–1.7) mm in size, and equipped with 37 collar spines on the head collar (dorsal spines in 2 alternating rows), being consistent with E. mekongi. In phylogenetic analyses, the adult flukes showed 99.0–100% homology based on cox1 sequences and 98.9–99.7% homology based on nad1 sequences with E. mekongi. The results evidenced that F. martensi cambodjensis snails act as the second intermediate host of E. mekongi, and hamsters can be used as a suitable experimental definitive host. As local people favor to eat undercooked snails, these snails seem to be an important source of human infection with E. mekongi in Cambodia.
-
Key words: Echinostoma mekongi, metacercaria, Filopaludina martensi cambodjensis snail, Pursat Province, Cambodia
INTRODUCTION
Among the 37-collar-spined
Echinostoma spp. or ‘
Echinostoma revolutum group’, total 26 valid or validity-retained spp. were acknowledged worldwide [
1]. They included 16 valid species, i.e.,
E. revolutum,
E. bolschewense,
E. caproni,
E. cinetorchis,
E. deserticum,
E. lindoense,
E. luisreyi,
E. mekongi,
E. miyagawai,
E. nasincovae,
E. novaezealandense,
E. paraensei,
E. paraulum,
E. robustum,
E. trivolvis, and
Echinostoma sp. IG of Georgieva et al., 2013, and 10 validity-retained species., i.e.,
E. acuticauda,
E. barbosai,
E. chloephagae,
E. echinatum,
E. jurini,
E. nudicaudatum,
E. parvocirrus,
E. pinnicaudatum,
E. ralli, and
E. rodriguesi [
1]. Recently, however,
E. robustum Yamaguti, 1935 has been put to a synonymy with
E. miyagawai Ishii, 1932 [
2]. Eight among the 26 species, including
E. revolutum,
E. cinetorchis,
E. echinatum,
E. lindoense,
E. mekongi,
E. miyagawai (experimental infection),
E. paraulum, and possibly
E. paraensei (from the coprolite of a human mummy), are regarded as human-infecting 37-collar-spined echinostomes [
1].
Echinostoma mekongi Cho, Jung, Chang, Sohn, Sinuon& Chai, 2020 (Digenea:
Echinostomatidae) was described as a new species based on adult flukes collected from 6 riparian people in Kratie and Takeo Province, Cambodia [
3]. This species was morphologically characterized by a small head collar, small collar spines, small oral and ventral suckers, and small cirrus sac compared to
E. revolutum or
E. miyagawai, and marked variation was noted in the morphology of testes, which are either globular or lobulated [
3]. However, its life history, including the first and second intermediate hosts and animal reservoir hosts as well as epidemiological characteristics, including geographical distribution, remained unknown. In the present study, we performed a life cycle study of
E. mekongi and discovered its metacercarial stage in a species (subspecies) of freshwater snail,
Filopaludina martensi cambodjensis, purchased from a local market in Pursat Province, Cambodia and obtained adult flukes through experimental infection to a hamster. The adult flukes were morphologically and molecularly (
cox1 and
nad1) confirmed to be
E. mekongi Cho et al., 2020 [
3].
MATERIALS AND METHODS
Ethical issue
To recover the adult flukes of E. mekongi, animal experiment was performed using a laboratory-bred Syrian golden hamster (Mesocricetus auratus). This animal experiment followed the guidelines of the Committee on the Ethics of Animal Experiments in Gyeongsang National University College of Medicine, Jinju, Korea (2017).
Collection of the metacercariae
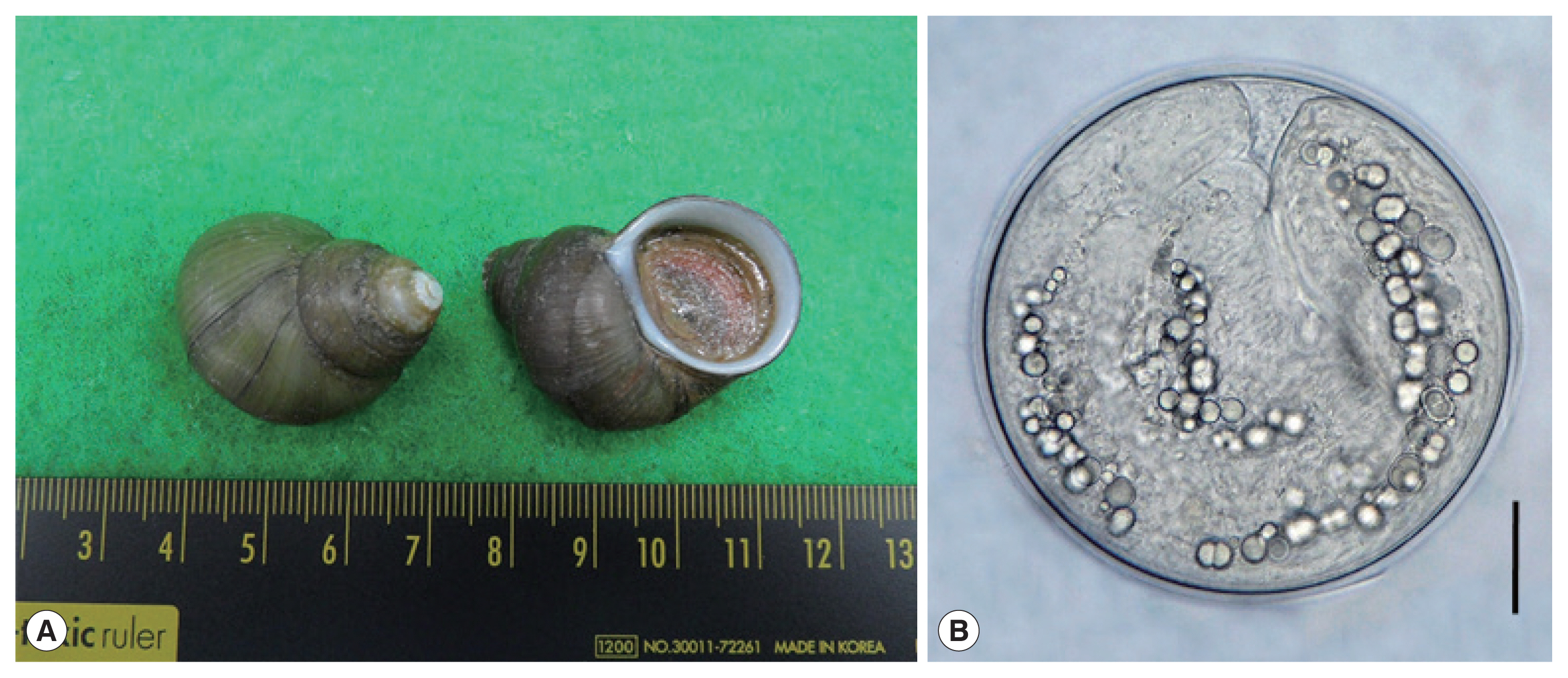
We purchased freshwater snails,
F. martensi cambodjensis (
Fig. 1A), from a local market in Pursat Province, Cambodia in May 2017. The identification of the snail species and subspecies was performed according to a previous report [
4]; in comparison with
Filopaludina martensi martensi, this subspecies is lacking in spiral ridges. The snails were transported to Korea (Department of Parasitology and Tropical Medicine, Gyeongsang National University College of Medicine, Jinju) under ice, and examined for echinostome metacercariae using the artificial digestion method as previously described [
5]. After crushing their shells with a mortar and pestle, they were digested with pepsin-HCl solution for 2 hr. Metacercariae were collected from the digested material under a stereomicroscope (Olympus, Tokyo, Japan).
The metacercariae were morphologically observed under a light microscope (Olympus). They (75 in number) were orally infected to an experimental hamster, and adult flukes were recovered in the small intestine at day 20 after the infection. The adult flukes were fixed with 10% neutral buffered formalin under a cover slip pressure, stained with Semichon’s acetocarmine, cleared in glycerin-alcohol, and mounted in glycerin-jelly for light microscopic observations. Some adult flukes were fixed in 70–80% ethanol for molecular analyses. To observe the surface ultrastructure, some adult flukes were washed several times with 0.2 M cacodylate buffer (pH 7.2), fixed with 2.5% glutaraldehyde at 4˚C, and washed 3 times with the buffer. They were then dehydrated through a graded ethanol series (50%, 70%, 80%, 90%, 95%, and absolute alcohol), dried with hexamethyldisilazane, coated (JFC-1100E ion sputtering device, Jeol, Tokyo, Japan) with gold, and observed using a scanning electron microscope (Jeol JSM-7610F, Tokyo, Japan) at an accelerating voltage of 15 kV.
Molecular analyses (cox1 and nad1)
For molecular analyses, worms preserved in ethanol were used. Genomic DNA was extracted using the Spin-Column Protocol of DNeasy® Blood&Tissue kit (Qiagen, Hilden, Germany). PCR was then conducted using specific primers designed to amplify
cox1 and
nad1 genes in echinostomes. The primers for
cox1 gene were JB3 and JB13, and the primers for
nad1 were JB11 and JB12 [
6]. The PCR products were purified and directly sequenced by Macrogen Inc. (Seoul, Korea). For evaluation of the genetic identity, the basic local alignment search tool (BLAST;
http://blast.ncbi.nlm.nih.gov/Blast.cgi) was applied. We used the Geneious® version 10.2.6 program (Biometers Ltd., Auckland, New Zealand) for alignment of the obtained sequences with GenBank reference
cox1 and
nad1 sequences of 37-collar-spined
Echinostoma spp. Multiple sequence alignment was performed using Clustal W program [
7]. The phylogenetic tree was constructed using the maximum-likelihood method based on Tamura-Nei model of nucleotide substitution with 1,000 bootstrap replications with our samples and several other 37-collar-spined
Echinostoma spp. deposited in GenBank, and viewed by MEGA X program [
8].
RESULTS
Recovery of metacercariae
A total of 115
E. mekongi metacercariae were detected in 60 (60.0%) out of 100
F. martensi cambodjensis snails examined (1–8 metacercariae per snail). The metacercariae (
Fig. 1B) were round and 174 (163–190) μm in diameter, having a thin cyst wall, 37 collar spines on the head collar, and characteristic excretory granules in 2 descending canals of the main excretory tube.
Twenty-eight adult flukes (37.3%; 28/75) were recovered from the small intestine of the hamster experimentally infected with 75 metacercariae at day 20 post-infection.
Adult flukes (
Fig. 2A–C) of
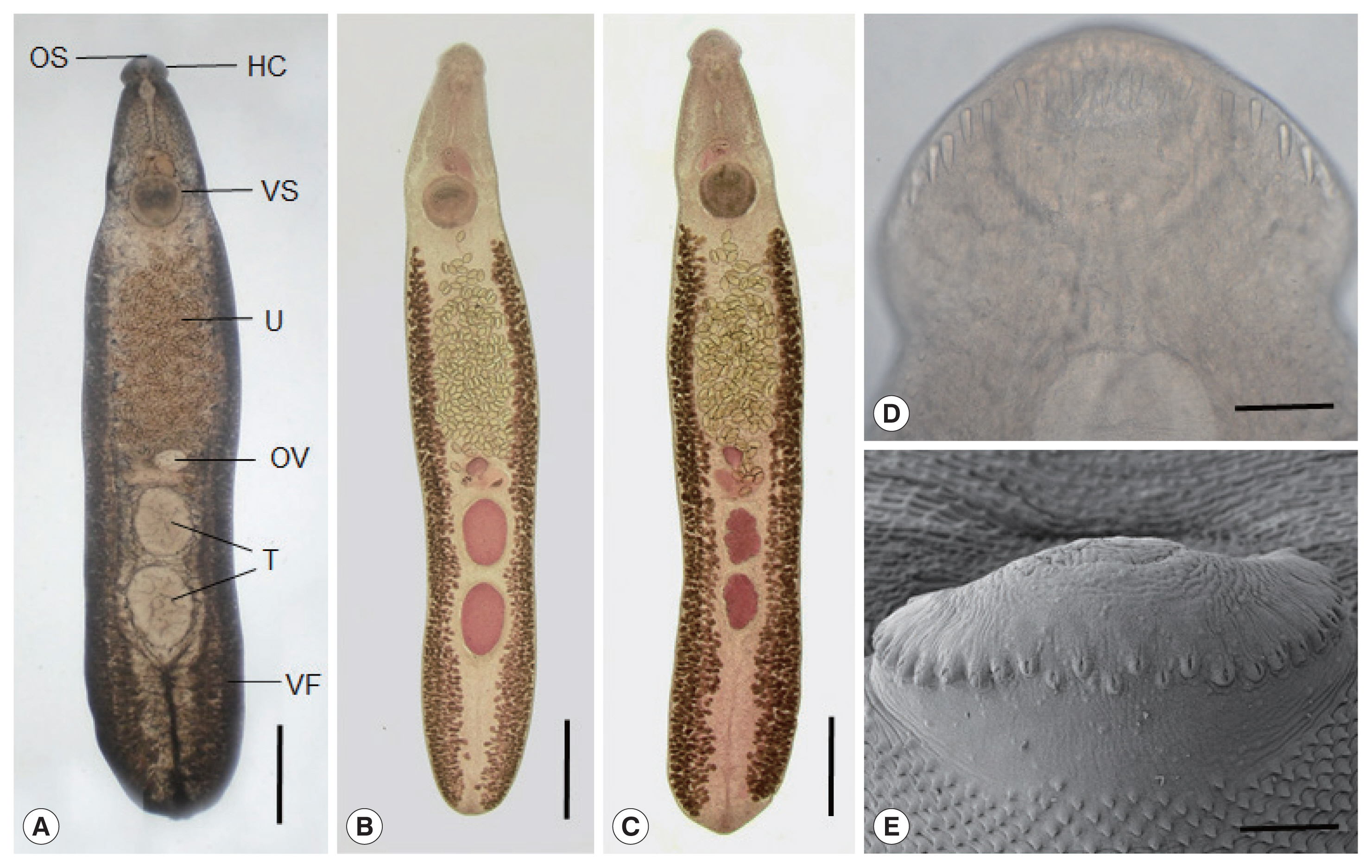
E. mekongi were elongated, ventrally curved, and 7.3 (6.4–8.2)×1.4 (1.1–1.7) mm in size (n=10). They revealed slight constriction of the body near the level of the ventral sucker. The head collar, 495 (445–525) μm in width (
Fig. 2D, E), was distinct, bearing 37 collar spines with 5 end group spines on each side, with the formula of 5-6-6-3-6-6-5. The oral sucker was subterminal, almost round, and 215 (200–230)×238 (220–255) μm. The prepharynx was short, 85 (70–95) μm in length, and the pharynx was muscular and well developed, 178 (150–190)×189 (160–210) μm. The esophagus was somewhat long, 448 (365–525) μm in length. The cirrus sac was well developed, 401 (380–420)×216 (205–230) μm, with a saccular seminal vesicle, and located between intestinal bifurcation and anterior margin of the ventral sucker. The ventral sucker was muscular and very large, 514 (460–550) ×555 (500–600) μm. The ovary was transversely elliptical, 197 (180–210)×242 (200–270) μm, and located on the median line of the body. Two testes were tandem, anterior testis 602 (470–720)×473 (330–730) μm, posterior testis 667 (470–750)×454 (310–580) μm, round, or slightly lobulated. Vitelline follicles were distributed in 2 lateral groups from the level of the posterior margin of the ventral sucker to near the posterior end of the body, not merging in the post-testicular field. Eggs were operculated, ovoid to elliptical, and 126 (108–133) ×75 (71–83) μm in size.
In SEM observations, the adult flukes were elongated, ventrally curved, had the largest width near the middle portion of the body, and equipped with 37 collar spines (figures not shown). A dorsal view of the head collar showed 2 alternating rows of dorsal collar spines (
Fig. 2E). Sensory papillae were distributed mainly on the tegument around the 2 suckers and head collar. Scale-like tegumental spines were densely distributed on the body surface (dorsal and ventral) between the head collar and ventral sucker. Their densities were decreased posteriorly on the ventral surface. The dorsal tegument near the excretory pore was devoid of spines.
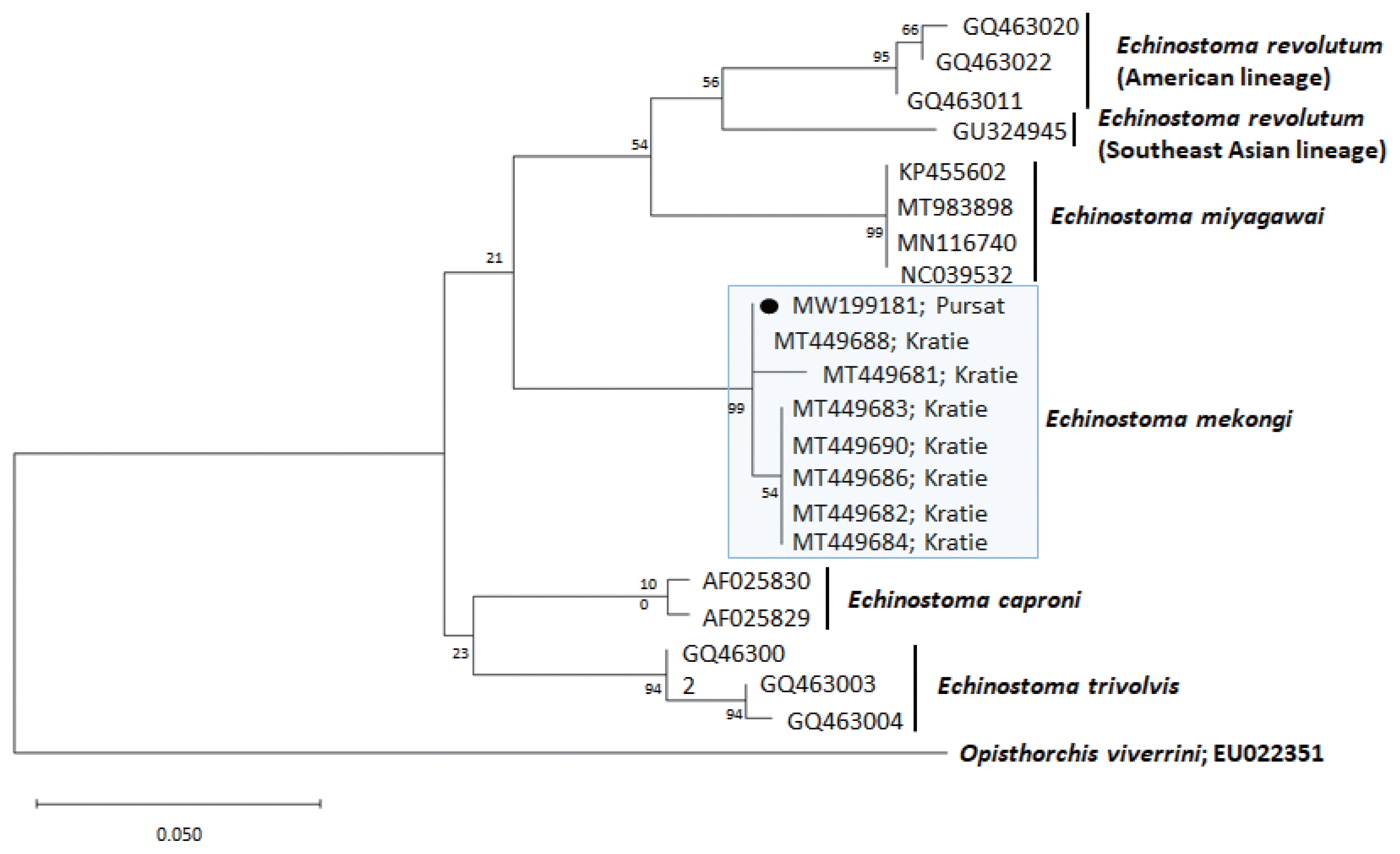
A phylogenetic tree based on
cox1 (247 bp after trimming) sequences was constructed using the ML analyses (
Fig. 3). The sequences of our sample (GenBank accession no. MW199181) clustered with high homology (99.0–100%) with the sequences of
E. mekongi deposited in GenBank, including isolates from Kratie Province, Cambodia (MT449681 and MT449688) (
Table 1). The homologies between our sample and the other 4
Echinostoma species were comparatively low (90.5–92.6%) (
Table 1).
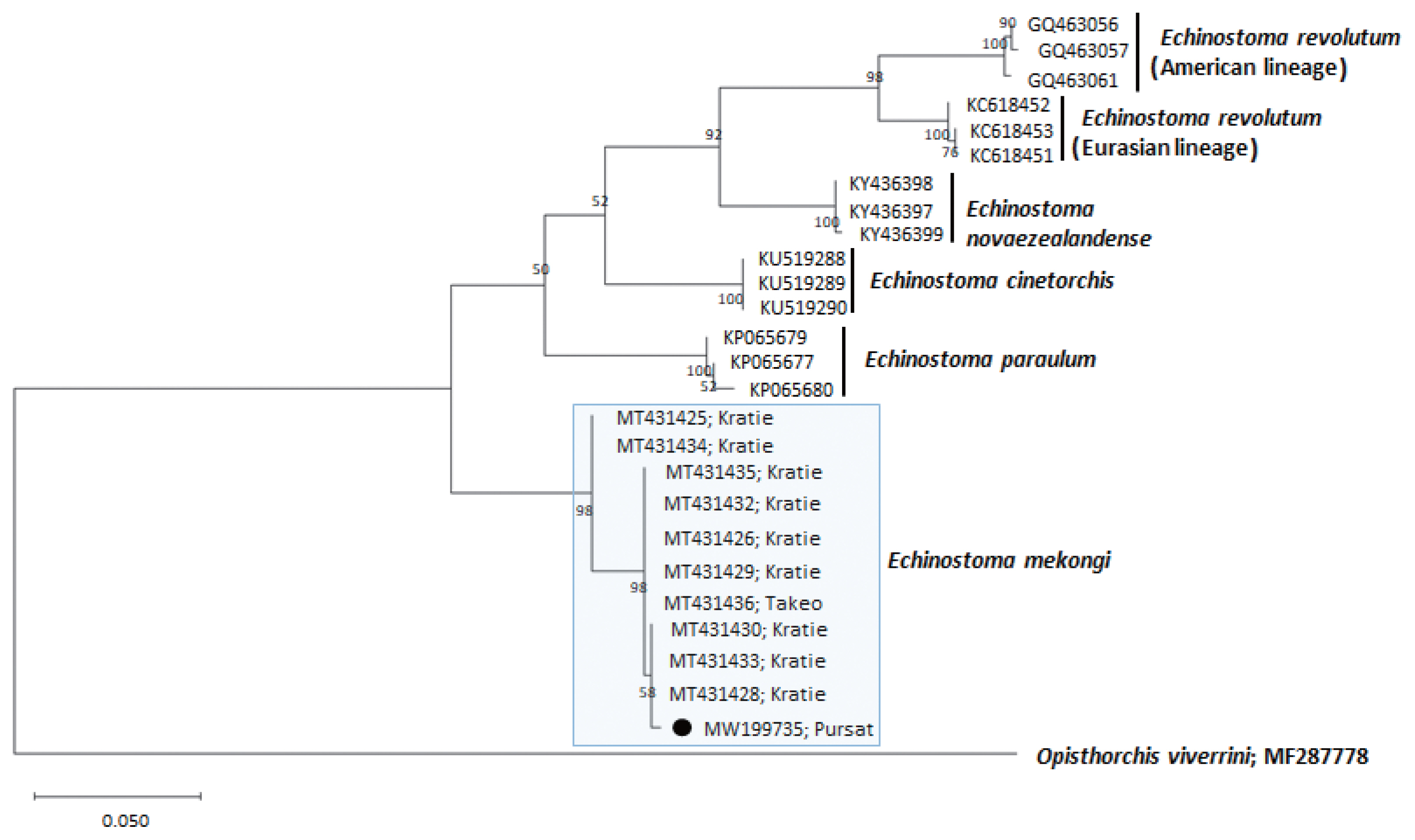
A phylogenetic tree based on
nad1 (348 bp after trimming) sequences was constructed also using the ML method (
Fig. 4). The sequences of our sample (MW119735) clustered with high homology (98.9–99.7%) with the sequences of
E. mekongi in GenBank, including isolates from Kratie Province, Cambodia (MT431430 and MT431425) (
Table 1). The homologies between our sample and the other 4
Echinostoma species were comparatively low (86.2–89.1%) (
Table 1).
DISCUSSION
The present study first confirmed that
F. martensi cambodjensis, a freshwater gastropod species, acts as a second intermediate host for
E. mekongi in Pursat Province, Cambodia. This snail species (
F. martensi) is widely distributed in Indochina Peninsula, including Vietnam, Thailand, and Cambodia [
4,
9–
11]. In Pursat Province, Cambodia, nearby the Tonle Sap Lake, these snails are commercially available in street markets together with larger snails, including
Pila spp. Local people favor to eat them undercooked. It is thus presumed that
F. martensi cambodjensis snails are a potential source of human infection with
E. mekongi. However, in Kratie and Takeo Provinces where human cases with
E. mekongi were detected [
3], the significance of these snails has not yet been verified, and it is urgently needed to examine these snails for
E. mekongi metacercariae.
The metacercariae of
E. mekongi were almost round (163–190 μm in diameter), equipped with 37 collar spines on the head collar, and morphologically indistinguishable from those of the other 37-collar-spined
Echinostoma spp. However, they were slightly or markedly larger than the metacercariae of
E. lindoense (120–130 μm),
E. revolutum (132–152 μm),
E. miyagawai (144–154 μm), and
E. trivolvis (135–170 μm) [
1].
It is referable that the same or closely related snail species, for example,
F. martensi martensi,
F. sumatrensis polygramma,
F. doliaris, and
Filopaludina sp., act as the second intermediate hosts of the other echinostome species, including
E. revolutum [
9–
11] and
Artyfechinostomum malayanum (syn.
Echinostoma malayanum) [
11]. It is also interesting to note that
F. martensi martensi snails can be used as a biological control agent of
Bithynia siamensis goniomphalos, the first intermediate host of
Opisthorchis viverrini [
12]. This may imply that these 2 trematode species are antagonistic and useful to understand epidemiology and geographical distribution of each species.
As
E. mekongi was originally reported from human infections [
3], and a hamster was susceptible for this echinostome infection in this study, a speculation could be made that
E. mekongi is an echinostome of mammalian hosts. However, many of the other 37-collar-spined
Echinostoma spp. are known to infect both birds and mammals, for example,
E. revolutum,
E. miyagawai, and
E. trivolvis [
1]. Although natural reservoir hosts for
E. mekongi have never been discovered, this may also be applied to
E. mekongi for taking both type of hosts. This point should be clarified by field surveys and also experimentally in the near future.
The first intermediate host of
E. mekongi is unknown. In
E. revolutum,
Lymnaea spp. and
Radix spp. are the most important snail species [
13], whereas, in
E. miyagawai,
Planorbis planorbis,
Anisus vortex, and
Radix peregra snails were reported to be natural first intermediate hosts [
14,
15]. Their cercariae are echinostome-type with an oral sucker armed with a spiny collar, ventral sucker, excretory bladder, and a moderately long tail [
13].
Regarding the morphology of
E. mekongi adults, several distinct characters described in the original report [
3] were reconfirmed in this study, although the size of the adult flukes were larger in specimens from human infections (av. 11.3 mm in length) than those from an experimental hamster in this study (av. 7.3 mm in length, 20-day-old). They morphologically resembled
E. revolutum,
E. miyagawai (syn.
E. robustum),
E. lindoense, and
E. trivolvis. They revealed slight body constriction near the level of the ventral sucker as reported in
E. miyagawai and
E. lindoense but not in
E. revolutum and
E. trivolvis [
1]. The adults of
E. mekongi had a smaller head collar, smaller collar spines, smaller suckers, and smaller cirrus sac compared to
E. revolutum and
E. miyagawai. They (
E. mekongi) also revealed variation in the morphology of testes; globular or slightly to markedly lobulated. Similar testis morphology was reported in
E. revolutum; their 2 testes are either entire or lobated [
16]. However, the testes morphology of
E. mekongi was comparable with that of
E. miyagawai and
E. lindoense; irregularly or deeply lobed (3–5 lobes) at times with horizontal extension in the former [
2] and deeply indented in the latter [
17]. Another unique feature of
E. mekongi adults was the shape of collar spines; they were not so long and not so sharply pointed as those of
E. revolutum,
E. miyagawai, and
E. trivolvis [
3]. In addition, the confluence of 2 lateral groups of vitelline follicles beyond the posterior testis field, which can be seen in
E. lindoense,
E. miyagawai, and
E. trivolvis [
1] was not recognized in
E. mekongi.
It should be referred that, in Pursat Province, Cambodia, human infections with
E. revolutum were previously reported [
18]. The cases were 4 schoolchildren who occasionally experienced vague abdominal pain and discomfort; they were fond of eating undercooked snails or clams of unidentified species caught near the Tonle Sap Lake and sold on the road to their homes after school [
18]. The worms recovered from the children were diagnosed as
E. revolutum based on morphological features [
18]. However, it is now strongly suggested that they might have been
E. mekongi. In particular, the figure in the paper shows a body constriction near the ventral sucker level in adult worms and relatively short and not sharply pointed collar spines on the head collar [
18] which is consistent with
E. mekongi rather than
E. revolutum. However, it is regretted that molecular analyses on the worms were not performed [
18]. Studies on human infections with
E. mekongi should be performed in Pursat Province and other localities of Cambodia as well as in different countries of Indochina Peninsula.
Notes
-
CONFLICT OF INTEREST
We have no conflict of interest related to this study.
ACKNOWLEDGMENTS
We are grateful to the staff of National Center for Parasitology, Entomology and Malaria Control, Phnom Penh, Cambodia, and the staff of Provincial Health Department, Pursat Province, Cambodia who helped this study.
Fig. 1(A) Filopaludina martensi cambodjensis snails purchased from a local market in Pursat Province, Cambodia. (B) A metacercaria of Echinostoma mekongi isolated from a F. martensi cambodjensis snail. Scale bar=30 μm.
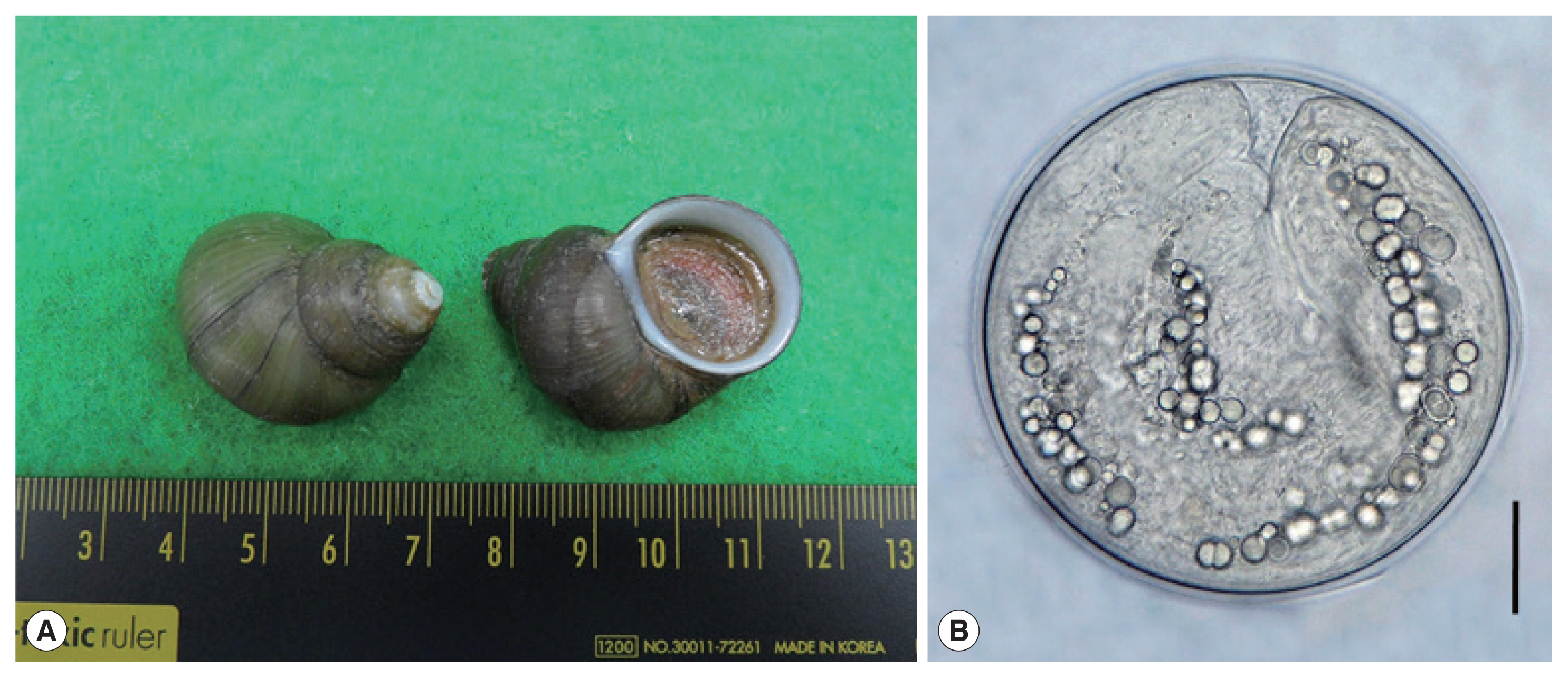
Fig. 2
Echinostoma mekongi adult worms obtained from an experimental hamster at day 20 post-infection. (A) An adult fluke showing the head collar (HC), oral (OS) and ventral suckers (VS), uterus (U) filled with eggs, ovary (OV), globular testes (T), and vitelline follicles (VF). Ventral view. Unstained. Scale bar=0.1 mm. (B, C) Other worms showing the same organs in Fig. 2A. Note the 2 globular (B) or slightly lobulated testes (C) and non-confluent vitellaria beyond the testicular field (B, C). Ventral view. Stained with Semichon’s acetocarmine followed by clearing in glycerin-alcohol and mounting in glycerin-jelly. Scale bar=0.1 mm (B, C). (D, E) Light microscopic (D) and scanning electron microscopic (E) views of the head collar with collar spines. Note the 2 alternating rows of the dorsal collar spines. Dorsal views. Scale bars=100 μm (D, E).
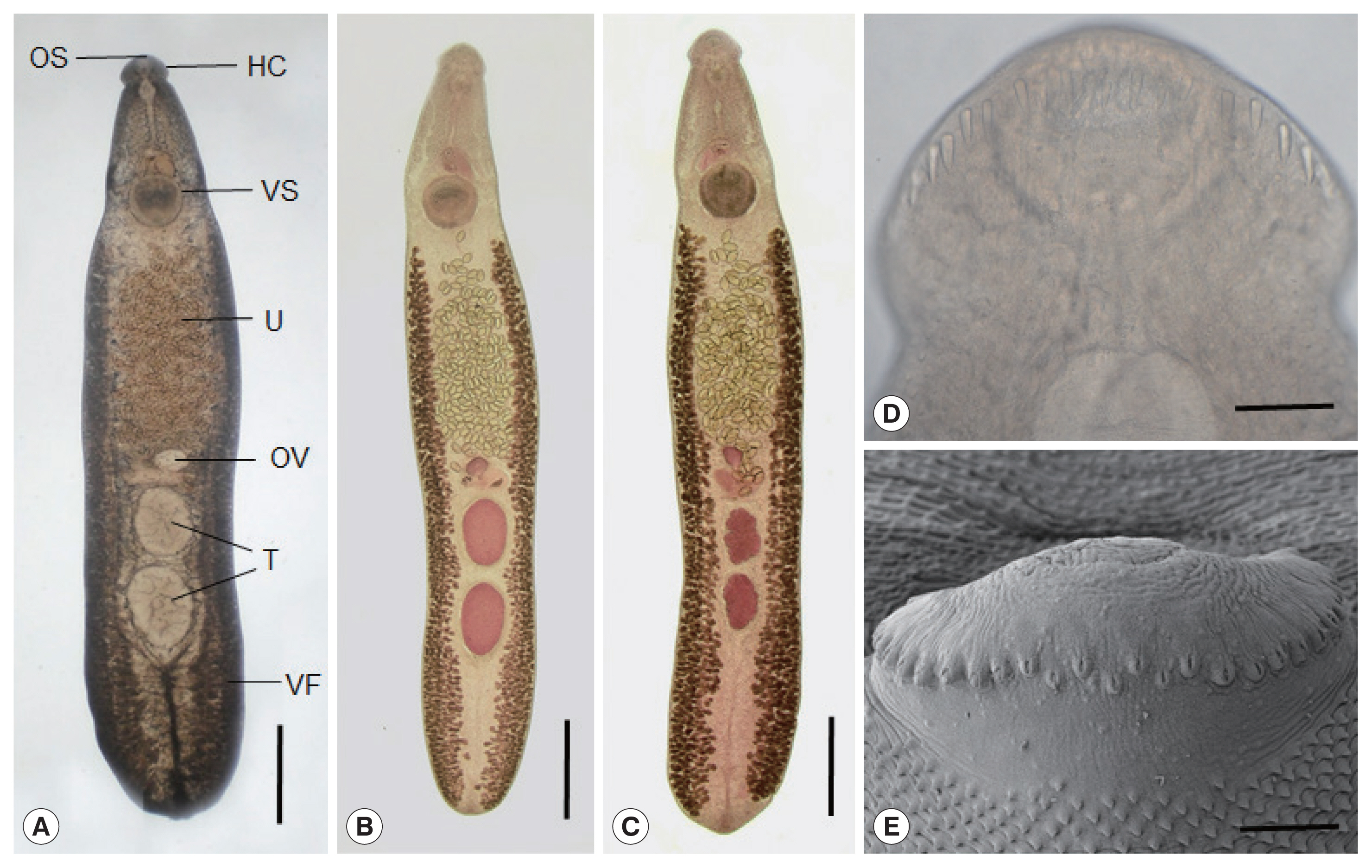
Fig. 3A phylogenetic tree of Echinostoma mekongi in comparison with 5 other 37-collar-spined Echinostoma spp. constructed based on 247 bp of mitochondrial cox1 sequences. Our sample (●, MW199181), from Pursat Province, is clustered with E. mekongi isolates from Kratie Province, Cambodia.
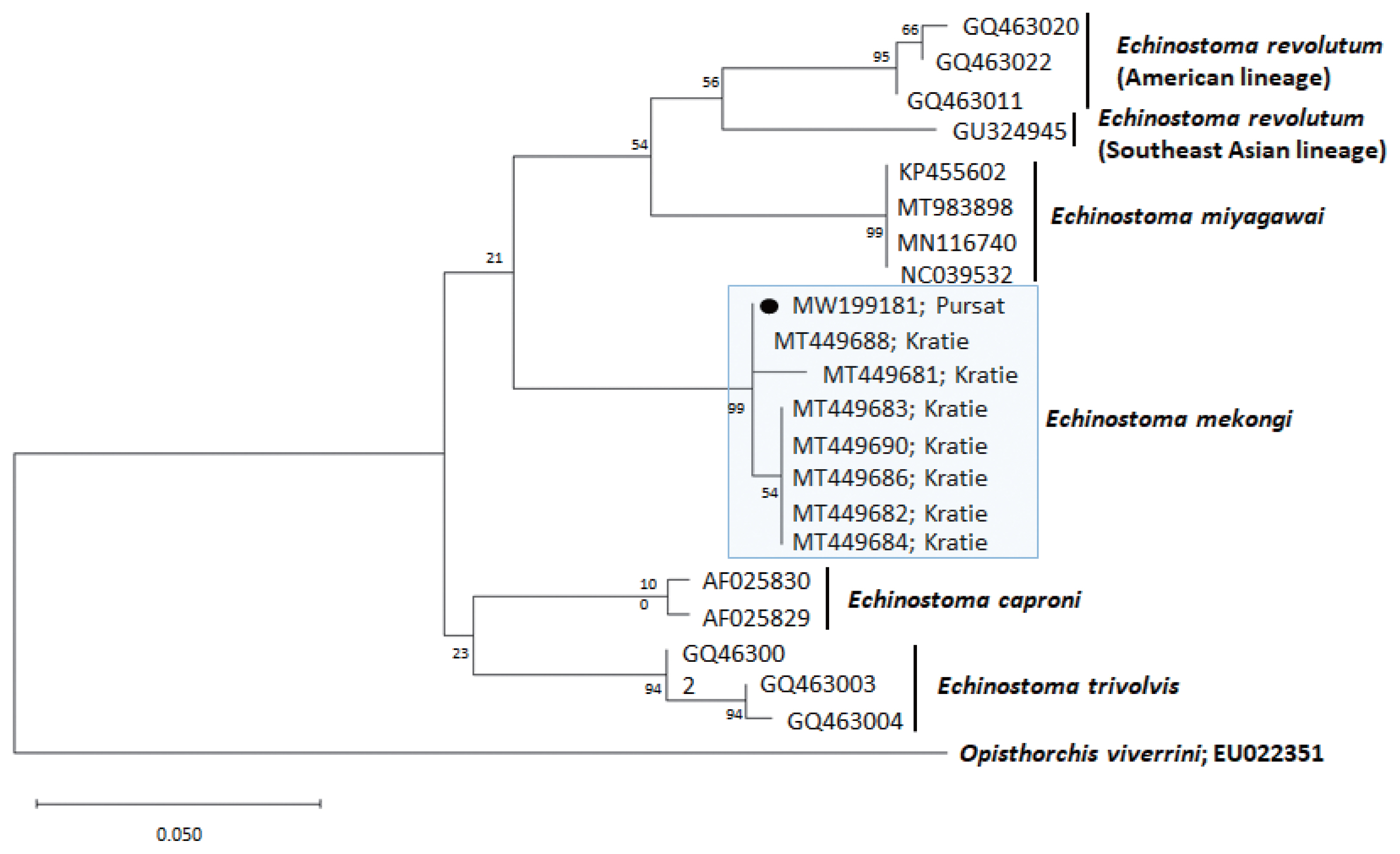
Fig. 4A phylogenetic tree of Echinostoma mekongi in comparison with 5 other 37-collar-spined Echinostoma spp. constructed based on 348 bp of mitochondrial nad1 sequences. Our sample (●, MW199735), from Pursat Province, is clustered with E. mekongi isolates from Kratie and Takeo Provinces, Cambodia.
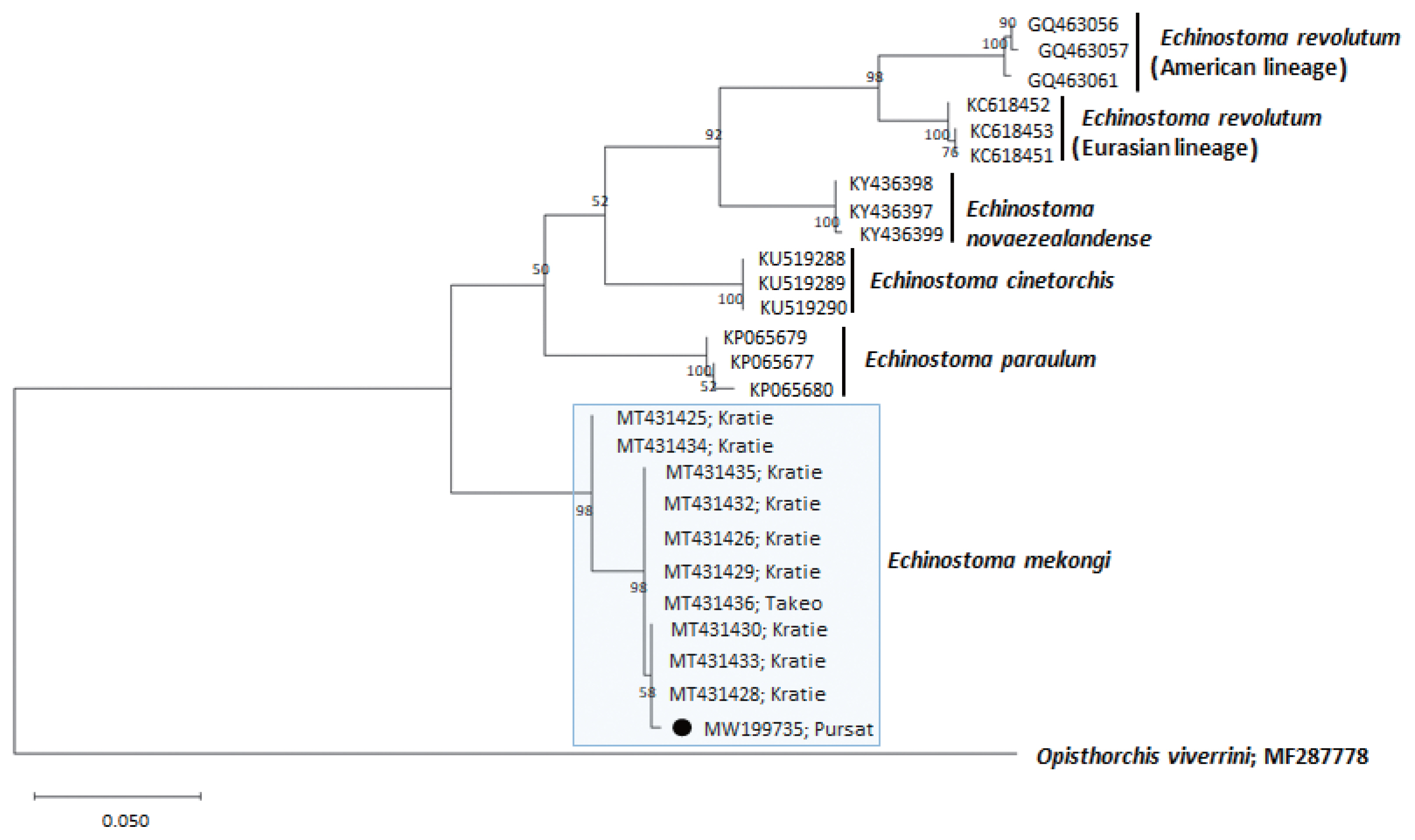
Table 1Comparison of our sample with Echinostoma mekongi from Kratie, Cambodia and other members of 37-collar-spined Echinostoma spp. based on cox1 and nad1 gene sequences
Table 1
|
cox1 |
nad1 |
|
|
|
Our samplea
|
|
Our samplea
|
|
Echinostoma mekongi (MT449688; Kratie Province) |
100.0 |
Echinostoma mekongi (MT431430; Kratie Province) |
99.7 |
|
|
Echinostoma mekongi (MT449681; Kratie Province) |
99.0 |
Echinostoma mekongi (MT431425; Kratie Province) |
98.9 |
|
|
Echinostoma caproni (AF025830) |
92.6 |
Echinostoma paraulum (KP065680) |
89.1 |
|
|
Echinostoma trivolvis (GQ463003) |
91.7 |
Echinostoma cinetorchis (KU519289) |
87.5 |
|
|
Echinostoma revolutum American lineage (GQ463020) |
91.7 |
Echinostoma revolutum American lineage (GQ463056) |
86.8 |
|
|
Echinostoma revolutum Southeast Asian lineage (GU324945) |
91.2 |
Echinostoma novaezealandense (KY436399) |
86.2 |
|
|
Echinostoma miyagawai (KP455602) |
90.5 |
Echinostoma revolutum Eurasian lineage (KC618453) |
86.2 |
References
- 1. Chai JY, Cho J, Chang T, Jung BK, Sohn WM. Taxonomy of Echinostoma revolutum and 37-collar-spined Echinostoma spp.: a historical review. Korean J Parasitol 2020;58:343-371.
https.//doi.org/10.3347/kjp.2020.58.4.343
- 2. Chai JY, Jung BK, Chang T, Shin H, Cho J, Ryu JY, Kim HS, Jeong MH, Hoang EH, Abdulla MBM.
Echinostoma miyagawai Ishii, 1932 (Echinostomatidae) from ducks in Aceh Province, Indonesia with special reference to its synonymy with Echimostoma robustum Yamaguti, 1935. Korean J Parasitol 2021;59:35-45.
https.//doi.org/10.3347/kjp.2021.59.1.35
- 3. Cho J, Jung BK, Chang T, Sohn WM, Sinuon M, Chai JY.
Echinostoma mekongi n. sp. (Digenea: Echinostomatidae) from riparian people along the Mekong River in Cambodia. Korean J Parasitol 2020;58:431-443.
https.//doi.org/10.3347/kjp.2020.58.4.431
- 4. Ng TH, Jeratthitikul E, Sutcharit C, Chhuoy S, Pin K, Pholyotha A, Siriwut W, Srisonchai R, Hogan ZS, Ngor PB. Annotated checklist of freshwater mollusk from the largest freshwater lake in Southeast Asia. ZooKeys 2020;958:107-141.
https.//doi.org/10.3897/zookeys.958.53865
- 5. Chai JY, Jung BK, Ryu JY, Kim HS, Hong SJ, Htoon TT, Tin HH, Na BK, Sohn WM. Larval gnathostomes and spargana in Chinese edible frogs, Hoplobatrachus rugulosus, from Myanmar: potential risk of human infection. Korean J Parasitol 58:467-473.
https.//doi.org/10.3347/kjp.2020.58.4.467
- 6. Morgan JA, Blair D. Relative merits of nuclear ribosomal internal transcribed spacers and mitochondrial CO1 and ND1 genes for distinguishing among Echinostoma species (Trematoda). Parasitology 1998;116:289-297.
https://doi.org/10.1017/S0031182097002217
- 7. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics 2007;23:2947-2948.
https://doi.org/10.1093/bioinformatics/btm404
- 8. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 2018;35:1547-1549.
https://doi.org/10.1093/molbev/msy096
- 9. Chai JY, Sohn WM, Na BK, De NV.
Echinostoma revolutum: metacercariae in Filopaludina snails from Nam Dinh Province, Vietnam, and adults from experimental hamsters. Korean J Parasitol 2011;49:449-455.
https.//doi.org/10.3347/kjp.2011.49.4.449
- 10. Chantima K, Chai JY, Wongsawad C.
Echinostoma revolutum: freshwater snails as the second intermediate hosts in Chiang Mai, Thailand. Korean J Parasitol 2013;51:183-189.
https.//doi.org/10.3347/kjp.2013.51.2.183
- 11. Noikong W, Wongsawad C, Chai JY, Saenphet S, Trudgett A. Molecular analysis of echinostome metacercariae from their second intermediate host found in a localised geographic region reveals genetic heterogeneity and possible cryptic speciation. PLoS Negl Trop Dis 2014;8:e2778.
https://doi.org/10.1371/journal.pntd.0002778
- 12. Wang YC, Liew TZ, Namsanor J, Sithithaworn P. Assessing the role of Filopaludina martensi martensi as a biocontrol agent of Bithynia siamensis goniomphalos, the first intermediate host of Opisthorchis viverrini
. Parasitol Res 2020;119:3415-3431.
https://doi.org/10.1007/s00436-020-06837-7
- 13. Chai JY. Chapter 2 Echinostomes Human Intestinal Fluke: From Discovery to Treatment and Control. Springer Nature; Dordrecht, The Netherlands. 2019, pp 1-549.
- 14. Kostadinova A, Gibson DI, Biserkov V, Chipev N. Re-validation of Echinostoma miyagawai Ishii, 1932 (Digenea: Echinostomatidae) on the basis of the experimental completion of its life-cycle. Syst Parasitol 2000;45:81-108.
https://doi.org/10.1023/A:1006241610689
- 15. Faltýnková A, Georgieva S, Soldánová M, Kostadinova A. A re-assessment of species diversity within the ‘revolutum’ group of Echinostoma Rudolphi, 1809 (Digenea: Echinostomatidae) in Europe. Syst Parasitol 2015;90:1-25.
https://doi.org/10.1007/s11230-014-9530-3
- 16. Kanev I. Life-cycle, delimitation and redescription of Echinostoma revolutum (Froelich, 1802) (Trematoda: Echinostomatidae). Syst Parasitol 1994;28:125-144.
https://doi.org/10.1007/BF00009591
- 17. Sandground JH, Bonne C.
Echinostoma lindoensis n. sp., a new parasite of man in the Celebes with an account of its life history and epidemiology. Am J Trop Med Hyg 1940;20:511-536.
https://doi.org/10.4269/ajtmh.1940.s1-20.511
- 18. Sohn WM, Chai JY, Yong TS, Eom KS, Yoon CH, Sinuon M, Socheat D, Lee SH.
Echinostoma revolutum infection in children, Pursat Province, Cambodia. Emerg Infect Dis 2011;17:117-119.
https.//doi.org/10.3201/eid1701.100920
 , Woon-Mok Sohn3,*
, Woon-Mok Sohn3,* , Jaeeun Cho1, Bong-Kwang Jung1
, Jaeeun Cho1, Bong-Kwang Jung1 , Taehee Chang1, Keon Hoon Lee1, Virak Khieu4, Rekol Huy4
, Taehee Chang1, Keon Hoon Lee1, Virak Khieu4, Rekol Huy4