Abstract
Legionella pneumophila is an opportunistic pathogen that survives and proliferates within protists such as Acanthamoeba spp. in environment. However, intracellular pathogenic endosymbiosis and its implications within Acanthamoeba spp. remain poorly understood. In this study, RNA sequencing analysis was used to investigate transcriptional changes in A. castellanii in response to L. pneumophila infection. Based on RNA sequencing data, we identified 1,211 upregulated genes and 1,131 downregulated genes in A. castellanii infected with L. pneumophila for 12 hr. After 24 hr, 1,321 upregulated genes and 1,379 downregulated genes were identified. Gene ontology (GO) analysis revealed that L. pneumophila endosymbiosis enhanced hydrolase activity, catalytic activity, and DNA binding while reducing oxidoreductase activity in the molecular function (MF) domain. In particular, multiple genes associated with the GO term ‘integral component of membrane’ were downregulated during endosymbiosis. The endosymbiont also induced differential expression of various methyltransferases and acetyltransferases in A. castellanii. Findings herein are may significantly contribute to understanding endosymbiosis of L. pneumophila within A. castellanii.
-
Key words: Acanthamoeba, Legionella, endosymbiosis, differential gene expression
Acanthamoeba spp. is one of the most abundant protozoan in the environment and commonly isolated from soil and water.
Acanthamoeba spp. trophozoite usually feeds on bacteria, fungi, algae or small organic particles by phagocytosis [
1]. However, some bacteria have developed strategies to resist phagocytosis, survive intracellularly and exploit
Acanthamoeba spp. for multiplication [
2]. These bacteria are able to survive in encysted
Acanthamoeba spp. which protects the endosymbionts from adverse environmental conditions [
3].
Acanthamoeba spp. not only enables the endosymbionts to persist in the environment but also enhances its pathogenicity [
4]. Moreover, since mammalian macrophages and amoebae show similar interactions with endosymbionts, investigating the endosymbiotic relationship between intracellular pathogens and
Acanthamoeba spp. would contribute to understanding how these organisms behave in the mammalian cells and its evasion of the human immune system.
Acanthamoeba spp. can be a host for a wide range of pathogenic microorganisms such as
Legionella pneumophila,
Chlamydophila pneumoniae,
Cryptococcus neoformans,
Mycobacterium avium,
Listeria monocytogenes, and
Pseudomonas aeruginosa,
etc [
3,
5–
7]. Among these microorganisms, the interaction between
Acanthamoeba spp. and
Legionella spp. is one of the most investigated. After uptake of
Legionella spp. by
Acanthamoeba spp.,
Legionella spp. forms a specialized compartment called
Legionella-containing vacuole (LCV). LCV avoids fusion with lysosomes to deter lysosomal digestion and also inhibits phagosomal maturation, thereby enabling
L. pneumophila to actively replicate inside the LCV [
8].
To date, LCV and a large number of effectors transferred by the intracellular multiplication/defective organelle transport (Icm/Dot) type IV system of
Legionella spp. have been identified [
9–
12]. Although the roles of these genes from
Legionella spp. have been evaluated, little research has been done on genes of
Acanthamoeba spp. during endosymbiosis with
Legionella spp. To understand the intracellular survival strategy of
Legionella spp., inhibition of phagosome lysis in
Acanthamoeba spp. needs to be studied. In this study, total transcriptional changes of
A. castellanii in response to survival and replication of
L. pneumophila during 12 hr and 24 hr were investigated by RNA sequencing analysis.
The LCV in the
Legionella-infected
A. polyphaga has been reported to remain intact for up to 8 hr post-infection (hr pi), disrupted by 12 hr pi, and eventually lysed to release the intracellular pathogens into the cytoplasm of the amoeba by 18 to 24 hr pi [
13].
L. pneumophila infection incurred the lysis of more than 80% of
A. polyphaga at 24 hr pi, and it has also been suggested that the intracellular condition may significantly differ between 12–18 hr pi and 24 hr pi [
13]. Contrary to the previous findings,
L. pneumophila-infected
A. castellanii in the present study remained intact even at 24 hr pi. Therefore, gene expression patterns at 12 hr pi and 24 hr pi were compared to confirm whether drastic differences were present in
L. pneumophila-infected
A. castellanii at these 2 time points.
A. castellanii was infected with
L. pneumophila [
14], and the
Legionella-infected
Acanthamoeba (L+A) was incubated for 12 hr and 24 hr at 25°C incubator. mRNA-Seq reads were mapped using TopHat software [
15], and differentially expressed gene were determined based on BEDtools and EdgeR [
16–
18]. And we used the FPKM (fragments per kilobase of exon per million fragments) as the method of determining the expression level of the gene regions. Gene classification was based on searches done by DAVID (
http://david.abcc.ncifcrf.gov/).
RNA samples from different experimental conditions were sequenced to investigate the endosymbiosis-induced gene expression changes in
A. castellanii (
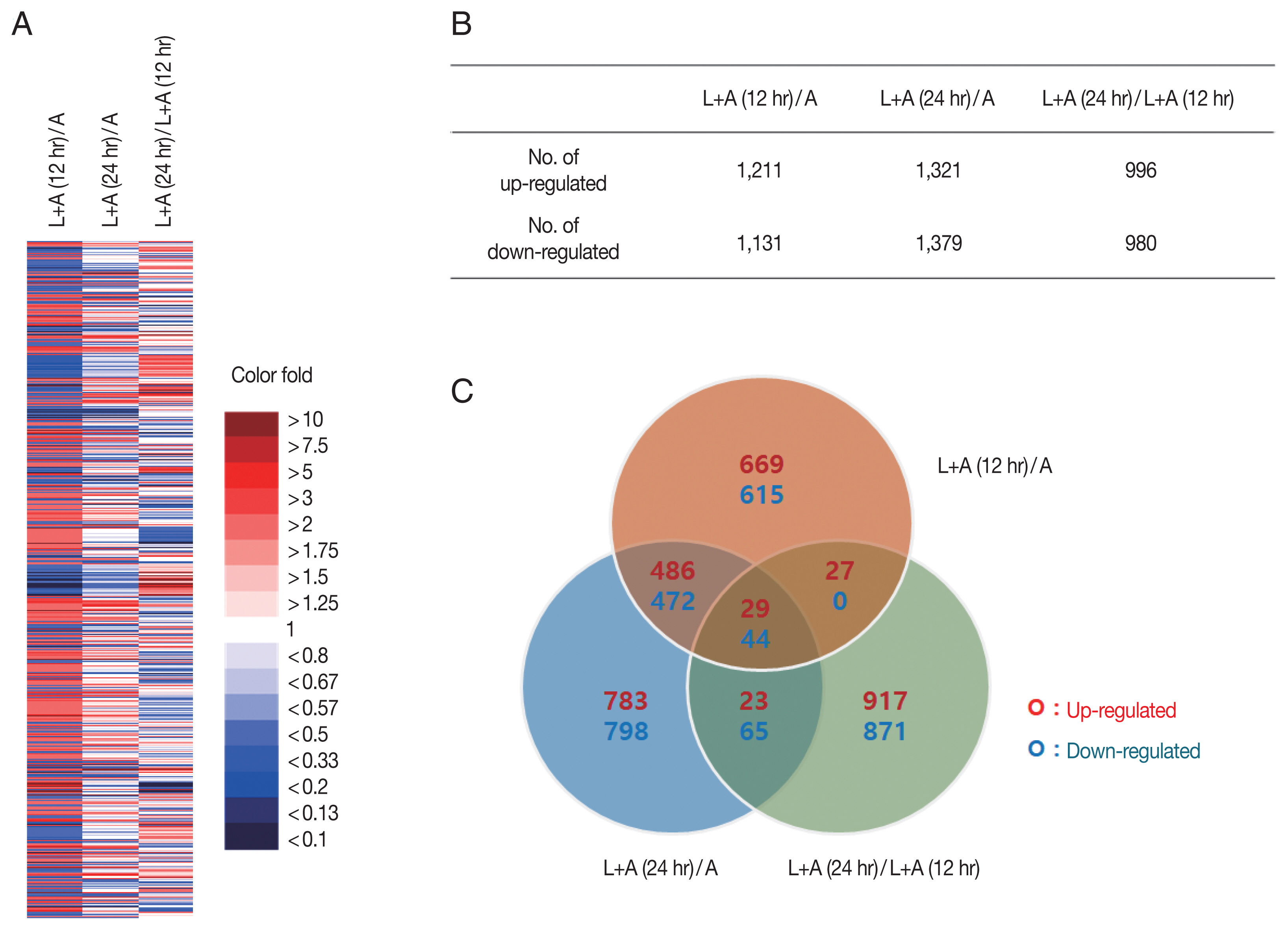
Fig. 1). A total of 7,108 genes whose expressions changed 12 hr pi and 24 hr pi were displayed using a heat map (
Fig. 1A). Genes from each group were colorized based on their expression level. Strongly upregulated/downregulated genes, as indicated by intense red/blue colors, were more prevalent in the 12 hr pi group than the 24 hr pi group. Genes whose expression levels changed more than 2 fold were selected for further analysis (
Fig. 1B). Among the 7,018 genes, 1,211 and 1,131 genes in the 12 hr pi group were upregulated and downregulated more than 2 fold, respectively. Similarly, 1,321 and 1,379 genes from the 24 hr pi group were upregulated and downregulated more than 2 fold, each respectively. Venn diagram revealed that a fraction of the DEGs found in the 12 hr pi overlapped with the DEGs from 24 hr pi group (
Fig. 1C). Our results revealed that 2,342 and 2,700 DEGs in
L. pneumophila-infected
A. castellanii at 12 hr pi and 24 hr pi were changed more than 2 fold, respectively. More DEGs were observed at 24 hr pi than at 12 hr pi, which may indicate that more gene involvement is required for survival of
L. pneumophila in the later stages of infection.
DEGs of
L. pneumophila-infected
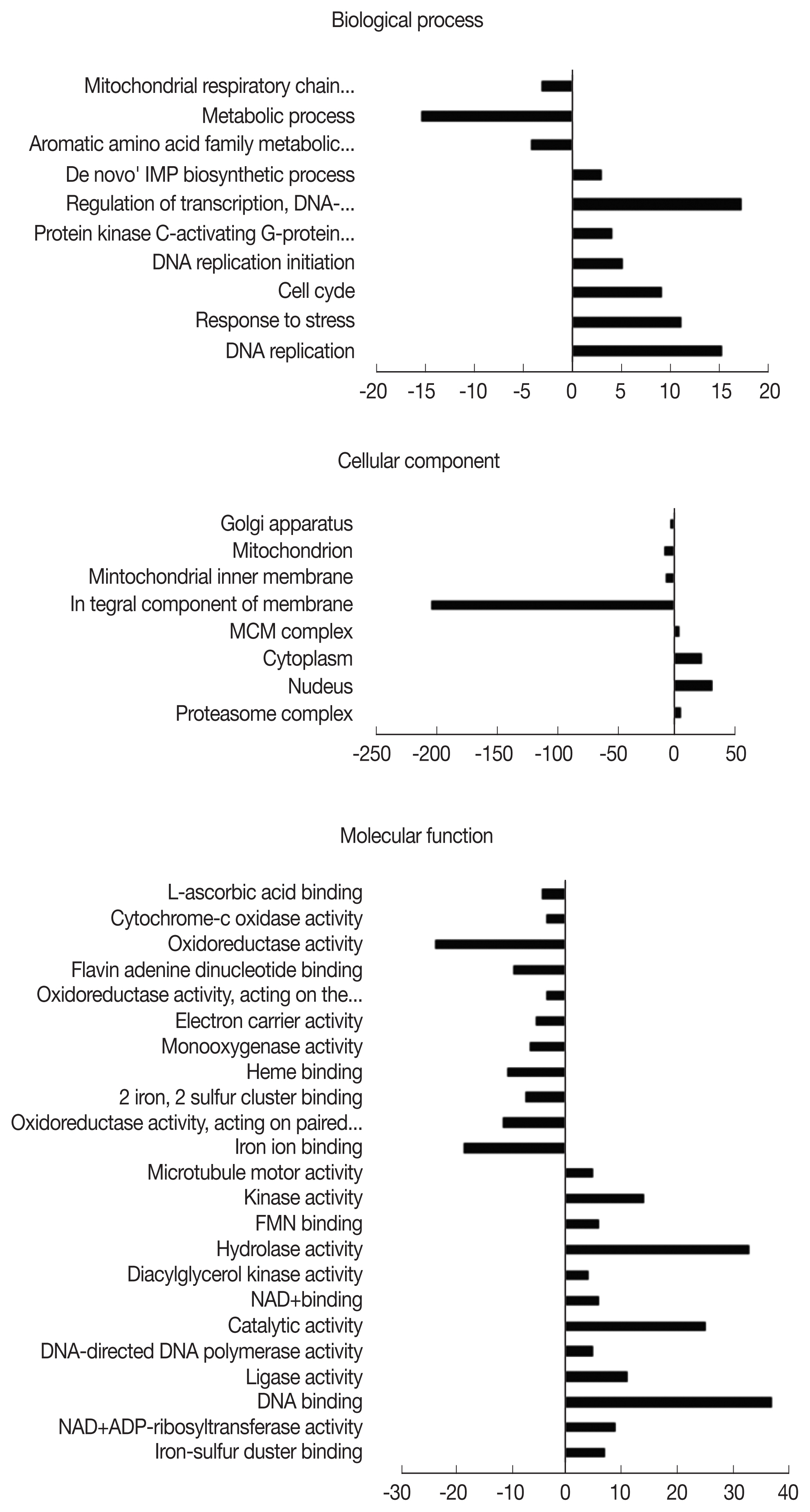
A. castellanii were assigned an Entrez Gene ID and subsequently classified into 3 domains: biological process (BP), cellular component (CC), and molecular function (MF). Classified DEGs were subdivided further into various gene ontology (GO) terms under each of the domains. In the 12 hr pi group, DEGs were assigned to 10 subcategories in BP, 8 subcategories in CC, and 23 subcategories in MF (
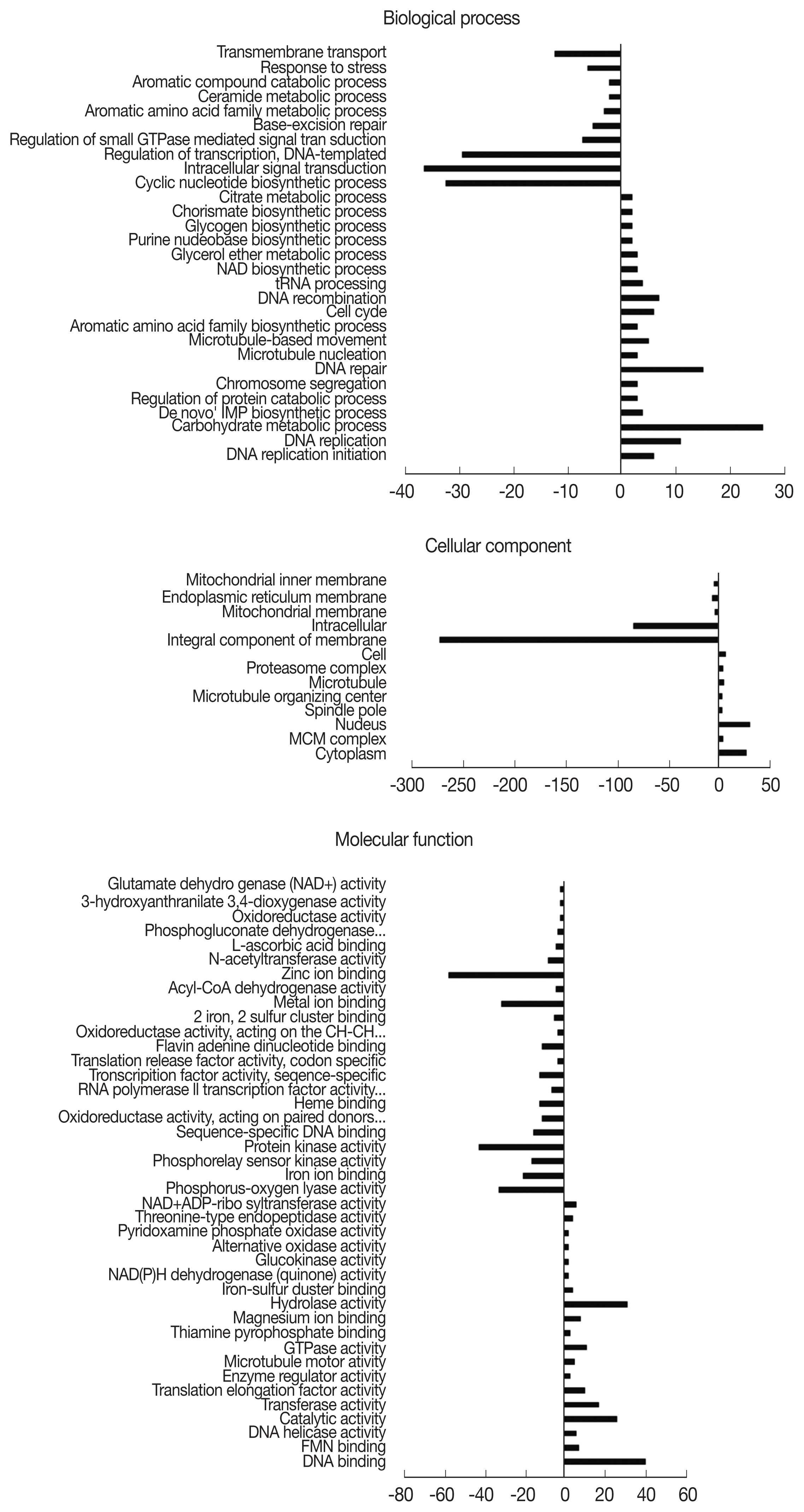
Fig. 2). During the 12 hr endosymbiosis, 17 genes in ‘regulation of transcription’ (domain: BP), 30 genes in the ‘nucleus’ (domain: CC), and 37 genes in ‘DNA binding’ (domain: MF) were determined to be the most upregulated genes. Within the CC domain, 200 downregulated genes were involved in the GO term ‘integral component of the membrane’. In the 24 hr pi group, DEGs were subdivided into 29, 13, and 41 GO terms under BP, CC, and MF domains, each respectively (
Fig. 3). In the BP domain, 26, 39, and 32 genes from the GO terms ‘regulation of transcription’, ‘intracellular signal transduction’, and ‘cyclic nucleotide biosynthetic process’ were drastically downregulated, each respectively. In the CC domain, similar to the 12 hr pi
A. castellanii, 270 downregulated genes were involved in the GO term ‘integral component of membrane’. In the MF domains, downregulated genes associated with the GO terms ‘zinc ion binding’ and ‘protein kinase activity’ were 57 and 42, respectively. From each of the domains, 26 genes from ‘carbohydrate metabolic process’ (domain: BP), 30 genes from the ‘nucleus’ (domain: CC), and 40 genes from ‘DNA binding’ (domain: MF) were mainly upregulated.
DEGs from
A. castellanii, which were upregulated or downregulated more than 10 fold post-infection with
L. pneumophila, were listed in
Tables 1 and
2. In the 12 hr pi
A. castellanii, 47 out of 1,211 DEGs were upregulated (
Table 1) and 90 out of 1,131 DEGs were downregulated more than 10 fold (
Table 2). Identities for several most upregulated proteins in this group were 2 hypothetical proteins (1,100 fold and 345 fold), S-adenosylmethionine-dependent methyltransferases (173 fold), and GDPD-mannose-3′, 5′-epimerase (87 fold) (
Table 1). GO analysis of the assigned Entrez Gene IDs revealed that the DEG which underwent 1,100 fold increase was a hypothetical protein that belonged to the DNA binding (GO: 0003677) category. Similarly, the GDPD-mannose-3′,5′-epimerase which was increased 87 fold, was associated with catalytic activity (GO: 0003824). Based on these findings, it can be speculated that these DEGs may be of importance during the initial phase of infection. While xylosyltransferase 1 was downregulated more than 700 fold, sulfiredoxin 1 was downregulated more than 500 fold, and vacuolar sorting-associated protein 13 were downregulated more than 12 fold. (
Table 2). GO analysis results revealed that the DEGs downregulated 10 fold or more were predominantly associated with the integral component of membrane (GO: 0016021). Findings are consistent with the changes in DEGs categorized under CC as illustrated in
Fig. 3.
Although 132 out of 1,321 DEGs were upregulated and 54 out of 1,379 DEGs were downregulated more than 10 fold in
A. castellanii 24 hr pi, approximately 60% of these DEGs (78 DEGs and 30 DEGs) were identified as hypothetical proteins. Strong inhibition of DEGs were observed in both 12 hr pi
A. castellanii (90 DEGs) and 24 hr pi
A. castellanii (53 DEGs). Among the DEGs demonstrating 10 fold or greater changes, 47 genes were upregulated while 90 genes were downregulated within the initial 12 hr pi (
Tables 1,
2). Conversely, by 24 hr pi, 132 upregulated and 54 downregulated DEGs were observed. From these results, we supposed that
L. pneumophila infection facilitated reduced
A. castellanii gene expression during the early stage of infection to inhibit phagocytic digestion, while enhancing the expression of specialized
A. castellanii genes during the late infection stage for LCV lysis and access to host cell machinery for intracellular replication.
Interestingly,
L. pneumophila-infected
A. castellanii showed differential expressions of methyltransferase-associated proteins. In addition to the S-adenosylmethionine-dependent methyltransferases and lysine methyltransferase enzyme domain-containing protein (
Table 1), 11 DEGs associated with methyltransferase were upregulated, and 19 DEGs were downregulated upon infection with
L. pneumophila for 12 hr. Furthermore,
L. pneumophila-infected
A. castellanii also demonstrated differential expressions of acetyltransferase-associated proteins. Histone acetyltransferase-associated protein was upregulated and 8 other acetyltransferases were downregulated 12 hr pi with
L. pneumophila. Icm/Dot type IV secretion system and its effectors of
L. pneumophila modulate host gene expression by altering the chromatin structure or by affecting the activities of transcription factors [
19]. Post-translational modifications such as DNA methylation, histone acetylation, and histone methylation have been shown to play a critical role in the epigenetic regulation of eukaryotic gene expression [
19,
20]. Our results revealed that
L. pneumophila-infected
A. castellanii showed differential expressions of 30 kinds of methyltransferase-associated proteins and 9 kinds of acetyltransferase-associated proteins at 12 hr pi. Based on the changes to epigenetic regulatory gene expressions, it can be speculated that
L. pneumophila can alter the gene expression of
A. castellanii through epigenetic mechanisms.
A plethora of DEGs induced in A. castellanii by the endosymbiont L. pneumophila were revealed in this study. However, 38.3% (1,930 of the 5,042) of A. castellanii genes were identified to be hypothetical proteins. Proportions of theses hypothetical proteins in the 12 and 24 hr pi groups can be ascribed to the lack of Acanthamoeba spp. database. Our investigation of the DEGs in A. castellanii by an endosymbiont provides important information to understanding the survival strategy utilized by notable intracellular pathogen L. pneumophila in A. castellanii. Future studies investigating the presence of an endosymbiosis-specific gene may help elucidate the underlying mechanism involved in L. pneumophila pathogenesis, which would contribute to understanding the inhibition of phagocytosis within A. castellanii or even immune evasion mechanism in human macrophages.
Notes
-
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
ACKNOWLEDGMENT
This work was supported by the National Research Foundation of Korea (NRF) grant funded by Korea government (MIST) (No. 2020R1F1A1068719).
Fig. 1An overview of significant changes in the gene expression profiles of A. castellanii. (A) Gene expressions under different experimental conditions displayed using a heat map. (B) The number of genes with significantly increased or decreased expression (more than 2 fold). (C) Venn diagram showing the number of overlapping genes differentially expressed among 3 experimental conditions. L+A(12 hr)/A; L. pneumohpila infected A. castellanii for 12 hr/A. castellanii, L+A(24 hr)/A; L. pneumohpila infected A. castellanii for 24 hr/A. castellanii, L+A(24 hr)/L+A(12 hr); L. pneumohpila infected A. castellanii for 24 hr/L. pneumohpila infected A. castellanii for 12 hr.

Fig. 2Distribution of gene ontology (GO) functional classifications. GO analysis of downregulated (left-hand direction) and upregulated (right-hand direction) genes in A. castellanii infected with L. pneumophila after 12 hr.

Fig. 3Distribution of gene ontology (GO) functional classifications. GO analysis of downregulated (left-hand direction) and upregulated (right-hand direction) genes in A. castellanii infected with L. pneumophila after 24 hr.

Table 1Genes upregulated more than 10 fold in A. castellanii 12 hr pi
Table 1
|
Gene symbol |
Fold change |
Annotation |
GO analysis |
|
|
|
|
L+A(12)/A |
L+A(24)/A |
Product |
Category (Term) |
|
ACA1_328910 |
1099.734 |
136.04 |
hypothetical protein |
MF (GO:0003677) |
|
|
ACA1_183610 |
345.332 |
0.920 |
hypothetical protein |
- |
|
|
ACA1_183700 |
173.774 |
0.462 |
S-adenosylmethionine-dependent methyltransferases |
- |
|
|
ACA1_140050 |
116.896 |
15.154 |
hypothetical protein |
CC (GO:0016021) |
|
|
ACA1_183570 |
87.745 |
0.232 |
GDPD-mannose-3′,5′-epimerase |
MF (GO:0003824) |
|
|
ACA1_324870 |
66.918 |
38.929 |
hypothetical protein |
- |
|
|
ACA1_300830 |
61.232 |
23.274 |
permeases of the major facilitator superfamily |
CC (GO:0016021) |
|
|
ACA1_139940 |
51.100 |
7.577 |
hypothetical protein |
- |
|
|
ACA1_183970 |
50.688 |
0.239 |
hypothetical protein |
CC (GO:0016021) |
|
|
ACA1_264780 |
41.374 |
2.313 |
hypothetical protein |
- |
|
|
ACA1_159010 |
36.741 |
11.383 |
NmrAlike family protein |
- |
|
|
ACA1_098380 |
27.710 |
0.838 |
hypothetical protein |
- |
|
|
ACA1_248200 |
25.965 |
3.365 |
phosphotransferase enzyme domain containing protein |
- |
|
|
ACA1_183940 |
25.733 |
0.905 |
GTPase activating Rap/RanGAP domainlike 3, putative |
- |
|
|
ACA1_096640 |
20.427 |
7.341 |
hypothetical protein |
- |
|
|
ACA1_183760 |
17.942 |
0.999 |
lysine methyltransferase enzyme domain containing protein |
- |
|
|
ACA1_376940 |
17.875 |
3.938 |
BNR/Aspbox repeat domain containing protein |
- |
|
|
ACA1_183580 |
17.602 |
0.724 |
S-adenosylmethionine-dependent methyltransferases |
- |
|
|
ACA1_270170 |
15.076 |
1.686 |
von Willebrand factor type A domain containing protein |
- |
|
|
ACA1_158840 |
15.058 |
6.332 |
metal dependent phosphohydrolase |
- |
|
|
ACA1_289630 |
14.765 |
11.367 |
hypothetical protein |
- |
|
|
ACA1_381540 |
14.601 |
0.503 |
hypothetical protein |
- |
|
|
ACA1_224160 |
14.371 |
6.761 |
Sec23/Sec24 beta-sandwich domain containing protein |
- |
|
|
ACA1_175370 |
14.029 |
6.388 |
Erf4 domain containing protein |
CC (GO:0016021) |
|
|
ACA1_140540 |
13.733 |
6.286 |
MORN repeatcontaining protein |
- |
|
|
ACA1_184710 |
13.687 |
26.561 |
Phospholipid methyltransferase domain containing protein |
CC (GO:0016021) |
|
|
ACA1_068540 |
13.266 |
5.034 |
Prokumamolisin, activation domain containing protein |
- |
|
|
ACA1_217750 |
13.172 |
4.030 |
phosphoenolpyruvate carboxykinase (GTP), putative |
MF (GO:0016301) |
|
|
ACA1_116700 |
13.149 |
9.620 |
hypothetical protein |
- |
|
|
ACA1_279740 |
12.960 |
5.501 |
hydrogenase assembly factor, putative |
MF (GO:0051536) |
|
|
ACA1_116690 |
12.553 |
5.917 |
hypothetical protein |
CC (GO:0016021) |
|
|
ACA1_285180 |
12.470 |
58.313 |
DNA breaking-rejoining enzyme domain containing protein |
MF (GO:0003677) |
|
|
ACA1_215790 |
12.170 |
8.113 |
copper/zinc superoxide dismutase |
- |
|
|
ACA1_275740 |
12.097 |
4.360 |
glycerol-3-phosphate dehydrogenase (soluble) |
MF (GO:0051287) |
|
|
ACA1_058320 |
12.094 |
4.914 |
GPR1/FUN34/yaaH family protein |
CC (GO:0016021) |
|
|
ACA1_358270 |
11.433 |
7.344 |
pyridine nucleotidedisulfide oxidoreductase domain containing protein |
MF (GO:0016491) |
|
|
ACA1_067720 |
11.300 |
8.046 |
hypothetical protein |
- |
|
|
ACA1_153710 |
11.001 |
0.845 |
RFX_DNA_binding |
BP (GO:0006355) |
|
|
ACA1_091110 |
10.950 |
0.842 |
hypothetical protein |
- |
|
|
ACA1_256560 |
10.936 |
3.671 |
hypothetical protein |
- |
|
|
ACA1_275730 |
10.900 |
2.743 |
phosphoglycerate mutase family domain containing protein |
- |
|
|
ACA1_165640 |
10.762 |
1.971 |
hypothetical protein |
- |
|
|
ACA1_060580 |
10.631 |
4.515 |
phosphatase |
- |
|
|
ACA1_245710 |
10.555 |
7.545 |
hypothetical protein |
- |
|
|
ACA1_325450 |
10.443 |
4.372 |
CBS domain containing protein |
- |
|
|
ACA1_187310 |
10.274 |
6.715 |
heme NO binding domain containing protein |
- |
|
|
ACA1_238590 |
10.109 |
3.454 |
CBS domain containing protein |
- |
Table 2Genes downregulated more than 10 fold in A. castellanii 12 hr pi
Table 2
|
Gene symbol |
Fold change |
Annotation |
GO analysis |
|
|
|
|
L+A(12)/A |
L+A(24)/A |
Product |
Category (Term) |
|
ACA1_113420 |
0.001 |
0.764 |
hypothetical protein |
- |
|
|
ACA1_112520 |
0.001 |
0.493 |
Cysteine-rich 4 helical bundle widely conserved |
- |
|
|
ACA1_111980 |
0.003 |
0.368 |
EF hand domain containing protein |
- |
|
|
ACA1_112090 |
0.003 |
0.454 |
hypothetical protein |
- |
|
|
ACA1_111740 |
0.007 |
0.190 |
hypothetical protein |
- |
|
|
ACA1_112480 |
0.007 |
0.492 |
Fbox domain containing protein |
- |
|
|
ACA1_376130 |
0.007 |
0.386 |
xylosyltransferase 1, putative |
- |
|
|
ACA1_058410 |
0.009 |
0.019 |
hypothetical protein |
- |
|
|
ACA1_147740 |
0.010 |
0.006 |
CBS domain containing protein |
- |
|
|
ACA1_113310 |
0.013 |
0.765 |
hypothetical protein |
- |
|
|
ACA1_166550 |
0.017 |
0.318 |
hypothetical protein |
- |
|
|
ACA1_374390 |
0.018 |
0.924 |
hypothetical protein |
- |
|
|
ACA1_101570 |
0.019 |
1.673 |
hypothetical protein |
- |
|
|
ACA1_392590 |
0.020 |
0.401 |
hypothetical protein |
- |
|
|
ACA1_060120 |
0.021 |
0.548 |
hypothetical protein |
- |
|
|
ACA1_400130 |
0.021 |
0.021 |
hypothetical protein |
- |
|
|
ACA1_307550 |
0.022 |
0.084 |
Fbox domain containing protein |
- |
|
|
ACA1_230230 |
0.022 |
0.022 |
hypothetical protein |
- |
|
|
ACA1_050390 |
0.024 |
2.233 |
3-oxoacyl-[acyl-carrier protein] reductase |
CC (GO: 0016021) |
|
|
ACA1_112110 |
0.024 |
0.463 |
glycosyl transferase |
CC (GO: 0016021) |
|
|
ACA1_112490 |
0.024 |
0.480 |
hypothetical protein |
- |
|
|
ACA1_063680 |
0.026 |
0.072 |
Reverse transcriptase |
- |
|
|
ACA1_390590 |
0.027 |
0.132 |
Hsp20/alpha crystallin superfamily protein |
- |
|
|
ACA1_063960 |
0.029 |
0.078 |
hypothetical protein |
- |
|
|
ACA1_112130 |
0.029 |
0.457 |
regulator of g protein signaling domain containing protein |
- |
|
|
ACA1_111970 |
0.029 |
0.399 |
sulfiredoxin 1 |
- |
|
|
ACA1_158820 |
0.029 |
0.646 |
hypothetical protein |
- |
|
|
ACA1_064370 |
0.029 |
0.029 |
AT Hook plus PHD finger transcription factor family member (athp1), putative |
- |
|
|
ACA1_064780 |
0.029 |
0.029 |
hypothetical protein |
- |
|
|
ACA1_064790 |
0.029 |
0.029 |
hypothetical protein |
- |
|
|
ACA1_340040 |
0.030 |
0.275 |
zinc finger, zz type domain containing protein |
- |
|
|
ACA1_350050 |
0.030 |
0.298 |
hypothetical protein |
- |
|
|
ACA1_112530 |
0.031 |
0.499 |
NLPC_P60 super family |
- |
|
|
ACA1_112180 |
0.032 |
0.527 |
major facilitator subfamily transporter |
CC (GO: 0016021) |
|
|
ACA1_112590 |
0.034 |
0.474 |
WH2 motif domain containing protein |
- |
|
|
ACA1_050380 |
0.036 |
1.885 |
hypothetical protein |
- |
|
|
ACA1_230220 |
0.037 |
0.025 |
hypothetical protein |
- |
|
|
ACA1_199000 |
0.038 |
0.229 |
SnoaL-like domain containing protein |
- |
|
|
ACA1_077210 |
0.040 |
0.376 |
hypothetical protein |
- |
|
|
ACA1_173000 |
0.043 |
0.352 |
Predicted NAD/FAD-dependent oxidoreductase |
- |
|
|
ACA1_326260 |
0.043 |
0.073 |
hypothetical protein |
- |
|
|
ACA1_060740 |
0.045 |
0.310 |
hypothetical protein |
- |
|
|
ACA1_270160 |
0.046 |
0.071 |
fascin subfamily protein |
- |
|
|
ACA1_200180 |
0.048 |
0.428 |
hypothetical protein |
- |
|
|
ACA1_383480 |
0.051 |
0.029 |
hypothetical protein |
- |
|
|
ACA1_155760 |
0.052 |
0.145 |
phosphoribosyltransferase |
- |
|
|
ACA1_207830 |
0.052 |
0.426 |
myotubularins and other putative membrane-associated proteins |
- |
|
|
ACA1_133180 |
0.052 |
0.254 |
Ser/Thr phosphatase family superfamily protein |
- |
|
|
ACA1_077290 |
0.053 |
0.638 |
5′nucleotidase |
CC (GO: 0016021) |
|
|
ACA1_365080 |
0.053 |
0.367 |
hypothetical protein |
- |
|
|
ACA1_383650 |
0.055 |
0.349 |
Human glyoxalase domain-containing protein 5 and similar proteins |
- |
|
|
ACA1_048480 |
0.056 |
0.616 |
hypothetical protein |
- |
|
|
ACA1_055330 |
0.059 |
0.292 |
N-terminal region of Chorein or VPS13 |
- |
|
|
ACA1_346470 |
0.060 |
0.450 |
RUN domain containing protein |
- |
|
|
ACA1_298420 |
0.062 |
0.056 |
hypothetical protein |
- |
|
|
ACA1_197730 |
0.064 |
0.823 |
hypothetical protein |
- |
|
|
ACA1_378930 |
0.064 |
0.329 |
hypothetical protein |
- |
|
|
ACA1_214630 |
0.067 |
0.147 |
hypothetical protein |
CC (GO: 0016021) |
|
|
ACA1_128200 |
0.068 |
0.051 |
CBS domain containing protein |
- |
|
|
ACA1_052800 |
0.071 |
0.571 |
O-methyltransferase family 3 protein |
- |
|
|
ACA1_112560 |
0.071 |
0.499 |
hypothetical protein |
- |
|
|
ACA1_322750 |
0.072 |
0.552 |
Glycosyl hydrolases family 2, TIM barrel domain |
- |
|
|
ACA1_383400 |
0.072 |
1.376 |
hypothetical protein |
CC (GO: 0016021) |
|
|
ACA1_253630 |
0.073 |
0.947 |
hypothetical protein |
- |
|
|
ACA1_391470 |
0.074 |
0.821 |
Hsp20/alpha crystallin superfamily protein |
- |
|
|
ACA1_066110 |
0.074 |
0.05 |
SCP-like extracellular protein domain containing protein |
- |
|
|
ACA1_131790 |
0.075 |
0.133 |
protein from patent family protein |
- |
|
|
ACA1_111930 |
0.075 |
0.311 |
carbonsulfur lyase, putative |
BP (GO: 0008152) |
|
|
ACA1_064380 |
0.077 |
0.052 |
hypothetical protein |
- |
|
|
ACA1_064940 |
0.077 |
0.029 |
Fbox domain containing protein |
- |
|
|
ACA1_323370 |
0.077 |
0.011 |
Hsp20/alpha crystallin superfamily protein |
- |
|
|
ACA1_383750 |
0.078 |
0.236 |
Ubiquitinconjugating enzyme subfamily protein |
- |
|
|
ACA1_006080 |
0.079 |
0.210 |
hypothetical protein |
CC (GO: 0016021) |
|
|
ACA1_180590 |
0.082 |
0.546 |
TRRAP family protein |
- |
|
|
ACA1_112980 |
0.083 |
0.755 |
protein kinase |
- |
|
|
ACA1_077300 |
0.083 |
0.247 |
serine/threonine kinase |
CC (GO: 0016021) |
|
|
ACA1_111880 |
0.084 |
0.225 |
Small acidic protein family |
- |
|
|
ACA1_372720 |
0.084 |
0.148 |
obtusifoliol 14alphademethylase, putative |
CC (GO: 0016021) |
|
|
ACA1_046720 |
0.086 |
0.076 |
Glycosyl hydrolase families |
- |
|
|
ACA1_324050 |
0.088 |
0.646 |
Vacuolar sorting-associated protein 13 [Intracellular trafficking and secretion] |
- |
|
|
ACA1_400540 |
0.088 |
0.076 |
sphingosine hydroxylase |
CC (GO: 0016021) |
|
|
ACA1_290200 |
0.089 |
0.329 |
hypothetical protein |
- |
|
|
ACA1_389110 |
0.09 |
1.016 |
hypothetical protein |
- |
|
|
ACA1_112500 |
0.094 |
0.535 |
TBC domain containing protein |
- |
|
|
ACA1_311650 |
0.095 |
0.096 |
hypothetical protein |
- |
|
|
ACA1_112060 |
0.096 |
0.429 |
O-methyltransferase, putative |
- |
|
|
ACA1_178260 |
0.097 |
0.118 |
cytochrome P450, putative |
MF (GO: 0005506) |
|
|
ACA1_066960 |
0.098 |
1.036 |
hypothetical protein |
- |
|
|
ACA1_046710 |
0.099 |
0.126 |
cytoplasmic protein, putative |
- |
|
|
ACA1_112640 |
0.100 |
0.498 |
MBOAT family protein |
CC (GO: 0016021) |
References
- 1. Visvesvara GS, Moura H, Schuster FL. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea
. FEMS Immunol Med Microbiol 2007;50:1-26.
https://doi.org/10.1111/j.1574-695X.2007.00232.x
- 2. Schmitz-Esser S, Toenshoff ER, Haider S, Heinz E, Hoenninger VM, Wagner M, Horn M. Diversity of bacterial endosymbionts of environmental Acanthamoeba isolates. Appl Environ Microbiol 2008;74:5822-5831.
https://doi.org/10.1128/AEM.01093-08
- 3. Greub G, Raoult D. Microorganisms resistant to free-living amoebae. Clin Microbiol Rev 2004;17:413-433.
https://doi.org/10.1128/cmr.17.2.413-433.2004
- 4. Richards AM, Von Dwingelo JE, Price CT, Abu Kwaik Y. Cellular microbiology and molecular ecology of Legionella-amoeba interaction. Virulence 2013;4:307-314.
https://doi.org/10.4161/viru.24290
- 5. Barker J, Brown MR. Trojan horses of the microbial world: protozoa and the survival of bacterial pathogens in the environment. Microbiology 1994;140:1253-1259.
https://doi.org/10.1099/00221287-140-6-1253
- 6. Essig A, Heinemann M, Simnacher U, Marre R. Infection of Acanthamoeba castellanii by Chlamydia pneumoniae
. Appl Environ Microbiol 1997;63:1396-1399.
https://doi.org/10.1128/AEM.63.4.1396-1399.1997
- 7. Guimaraes AJ, Gomes KX, Cortines JR, Peralta JM, Peralta RH.
Acanthamoeba spp. as a universal host for pathogenic microorganisms: One bridge from environment to host virulence. Microbiol Res 2016;193:30-38.
https://doi.org/10.1016/j.micres.2016.08.001
- 8. Isberg RR, O’Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: making a cosy niche inside host cells. Nat Rev Microbiol 2009;7:13-24.
https://doi.org/10.1038/nrmicro1967
- 9. Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila
. Science 1998;279:873-876.
https://doi.org/10.1126/science.279.5352.873
- 10. Steiner B, Weber S, Hilbi H. Formation of the Legionella-containing vacuole: phosphoinositide conversion, GTPase modulation and ER dynamics. Int J Med Microbiol 2018;308:49-57.
https://doi.org/10.1016/j.ijmm.2017.08.004
- 11. Cazalet C, Rusniok C, Brüggemann H, Zidane N, Magnier A, Ma L, Tichit M, Jarraud S, Bouchier C, Vandenesch F, Kunst F, Etienne J, Glaser P, Buchrieser C. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet 2004;36:1165-1173.
https://doi.org/10.1038/ng1447
- 12. Cazalet C, Gomez-Valero L, Rusniok C, Lomma M, Dervins-Ravault D, Newton HJ, Sansom FM, Jarraud S, Zidane N, Ma L, Bouchier C, Etienne J, Hartland EL, Buchrieser C. Analysis of the Legionella longbeachae genome and transcriptome uncovers unique strategies to cause Legionnaires' disease. PLoS Genet 2010;6:e1000851.
https://doi.org/10.1371/journal.pgen.1000851
- 13. Molmeret M, Bitar DM, Han L, Kwaik YA. Disruption of the phagosomal membrane and egress of Legionella pneumophila into the cytoplasm during the last stages of intracellular infection of macrophages and Acanthamoeba polyphaga
. Infect Immun 2004;72:4040-4051.
https://doi.org/10.1128/IAI.72.7.4040-4051.2004
- 14. Mou Q, Leung PHM. Differential expression of virulence genes in Legionella pneumophila growing in Acanthamoeba and human monocytes. Virulence 2018;9:185-196.
https://doi.org/10.1080/21505594.2017.1373925
- 15. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25:1105-1111.
https://doi.org/10.1093/bioinformatics/btp120
- 16. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010;26:841-842.
https://doi.org/10.1093/bioinformatics/btq033
- 17. Varet H, Brillet-Guéguen L, Coppée JY, Dillies MA. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS One 2016;11:e0157022.
https://doi.org/10.1371/journal.pone.0157022
- 18. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004;5:R80.
https://doi.org/10.1186/gb-2004-5-10-r80
- 19. Qiu J, Luo ZQ.
Legionella and Coxiella effectors: strength in diversity and activity. Nat Rev Microbiol 2017;15:591-605.
https://doi.org/10.1038/nrmicro.2017.67
- 20. Yen CY, Huang HW, Shu CW, Hou MF, Yuan SS, Wang HR, Chang YT, Farooqi AA, Tang JY, Chang HW. DNA methylation, histone acetylation and methylation of epigenetic modifications as a therapeutic approach for cancers. Cancer Lett 2016;373:185-192.
https://doi.org/10.1016/j.canlet.2016.01.036