Abstract
Babesia microti is one of the most common causative agents of babesiosis. A sensitive and rapid detection is necessary for screening potentially infected individuals. In this study, B. microti cytochrome c oxidase subunit I (cox1) was selected as the target gene, multiple primers were designed, and optimized by a recombinase-aided amplification (RAA) assay. The optimal primers and probe were labeled with fluorescein. The sensitivity of fluorescent RAA (fRAA) was evaluated using gradient diluents of the cox1 recombinant plasmid and genomic DNA extracted from whole blood of B. microti infected mice. The specificity of fRAA was assessed by other transfusion transmitted parasites. The analytical sensitivity of the fRAA assay was 10 copies of recombinant plasmid per reaction and 10 fg/μl B. microti genomic DNA. No cross-reaction with any other blood-transmitted parasites was observed. Our results demonstrated that the fRAA assay would be rapid, sensitive, and specific for the detection of B. microti.
-
Key words: Babesia microti, recombinase-aided amplification, molecular detection
Human babesiosis is an emerging tick-borne zoonotic disease and a new global threat affecting human health. It is caused by infection with intraerythrocytic parasites of the genus
Babesia [
1]. Healthy people who naturally infected are always asymptomatic, and even immunocompetent patients are mild or subclinical. However, low-grade or asymptomatic parasitemia may persist for years [
2]. Babesia parasites are transmitted to humans and animals by ixodid ticks.
Babesia microti has become a high-risk pathogen that is transmitted by blood transfusion, particularly in the northeast and upper mid-west of the United States [
3]. The prevalence of
Babesia spp. in China is currently underestimated due to the lack of epidemiological data. Sensitive screening and diagnostic methods are needed to define the impact of this parasite. Multiple methodologies have been used to detect
B. microti. Microscopic examination, antibody/antigen assays, and nucleic acid tests (NAT) exhibited different sensitivity and specificity during the different stages of
B. microti infection [
4]. Although microscopic examination, as the “gold standard” method, has widely been used to confirm the parasitic infection, its low sensitivity limits its use in blood donors with low levels of parasitemia [
5]. The indirect immunofluorescence assay (IFA) is the most widely used serological method, but IFAs differ in which antibody class or classes are detected and in cutoff titers are determined by hand [
6]. Furthermore, IFAs fail to distinguish between present (active) and past (resolved) infections. The application of NAT technology, including polymerase chain reaction (PCR) [
7] and transcription-mediated amplification (TMA) [
8] for blood screening has dramatically reduced the risk of transfusion infection worldwide [
9]. However, the cost-effectiveness of NAT, particularly in resource-constrained countries, limits the implementation. Even in the United States, where
B. micrioti infection is the major cause of transfusion-transmitted babesiosis, NAT is selectively implemented only in epidemic areas [
10].
It is therefore necessary to develop cheaper molecular screening methods as an alternative to PCR. Several detection systems have been developed and used for routine detection, including loop-mediated isothermal amplification (LAMP) [
11] and recombinase polymerase amplification (RPA) [
12]. Recombinase-aided amplification (RAA) is a novel nucleic acid isothermal amplification technology. The basic RAA system includes recombinase UvsX, single-stranded DNA binding protein, and a DNA polymerase (all from
E. coli). With high sensitivity, specificity, and simplicity, it can rapidly detect nucleic acids under non-laboratory conditions. Thus, RAA has widely been used in the detection of several pathogens and the amplified products can be detected by agarose gel electrophoresis, a real-time fluorescence detection platform or a lateral flow strip (LFS). In the current study, we established and evaluated a rapid and simple fluorescence RAA (fRAA) assay for detecting
B. microti infection.
Many molecular assays are used to screen and differentiate parasites infections and they always amplify the 18S rRNA gene, which allows for detection of a broad-range of
Babesia spp. The mitochondrial genome in apicomplexan parasites, including
Babesia,
Theileria,
Plasmodium spp., and
Toxoplasma gondii, has similar structure [
13]. Among the 4 distinct mitochondrial genes, cytochrome oxidase
c subunit I (
cox1) is evolutionarily conserved and presents in a larger copy number than the chromosomal genes [
14]. The sequences of
B. microti cox1 gene in NCBI are highly homologous (100%) by MegAlign in Lasergene 7.1 (DNAStar, Madison, Wisconsin, USA) among
B. microti strains but different from other apicomplexan parasites, making it a valuable target for molecular detection.
The recombinant plasmid containing a 500 bp fragment of the
cox1 gene (GenBank No. LC005813.1) (nt 263–762) were synthesized, cloned to the pUC57 vector (designated on pUC57-
cox1), and used as the standard positive template for fRAA assay. Several forward and reverse RAA primers were designed (
Supplementary Table S1) and different primer pairs were optimized by a commercial RAA kit (Qitian, Wuxi, China). The amplified products ranging from 170 to 239 bp were screened using 1.5% agarose gel electrophoresis. After screening, the best primer pairs were chosen (
Supplementary Fig. S1). The forward primer was labeled with FAM at the 5′-end (5′-FAM-CTTGGTCTATCTATATAACATCTGTGTTATTG-3′) and the reverse primer was labeled with BHQ-1 at the 3′-end (5′-CTGGATGTCCAAAGACCCAGAATAGATGCCGAT-BHQ-1–3′). The probe was designed by adding a fluorescence FAM on the 31st base (T) from the 5′-end, an internal tetrahydrofuran residue (THF) on the 32nd base (C), BHQ1 on the 33rd base (T), and a C3 spacer (SpC3) at the 3′-end (5′-CATGCTTCTTGCTGATAGGCACTATAACAC [FAM-dT] C [THF] T [BHQ1-dT] GCTATTTG ATCCTAC-3′ SpC3). All primers and probe used for fRAA assay were synthesized by Sangon Biotech (Shanghai, China).
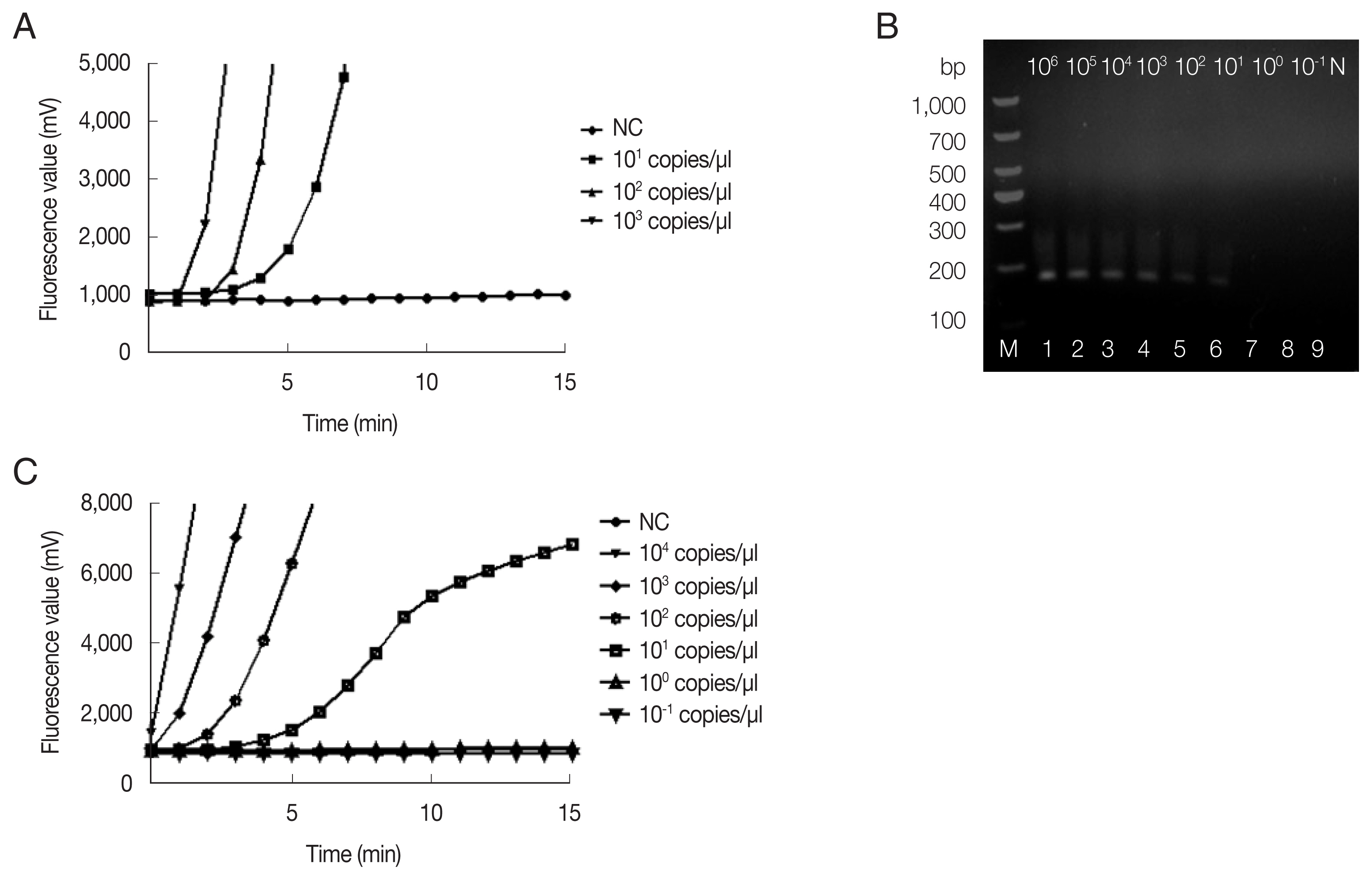
The fRAA assays were performed in 50 μl using a commercial fluorescence RAA kit according to the manual supplied by the manufacturer (Qitian, Wuxi, China). The reaction mixtures contained 2 μl of plasmid DNA, 25 μl of reaction buffer, 15.7 μl of DNase-free distilled water, 2.1 μl of primer F (10 μM), 2.1 μl of primer R (10 μM), 0.6 μl of the probe (10 μM), and 2.5 μl of 280 mM magnesium acetate. The reaction mixture was added to a tube containing the RAA enzyme mix (SSB, 800 ng/μl; UvsX, 120 ng/μl; DNA polymerase, 30 ng/μl) in a lyophilized form. The tubes were then transferred to a QT-F7200-0001 fluorescence detector (Qitian) at 37°C for 20 min. The fRAA developed could detect 10 copies/μl recombinant plasmids in each experiment (
Fig. 1A). Approximately 100 copies of 18S rRNA gene in a real-time PCR can be reliably detected [
7].
Copy number analysis using southern hybridization estimated approximately 20 copies of the mitochondrial genome per haploid nuclear genome of
B. microti [
14], which suggested the limit of detection (LOD) was about 0.5 parasite/μl. Thus, the assay was up to 200-fold more sensitive than Giemsa-stained blood smear, which was estimated to be 10–50 parasites/μl under ideal conditions, but about 100 parasites/μl for routine diagnostic screening [
16]. Our result showed that the fRAA assay had favorable sensitivity. In a recent study, LFD-RPA exhibited a slightly higher sensitivity of 0.25 parasite/μl blood [
12].
Blood from the
B. microti infected (ATCC, strain PRA-99) BALB/c mice (IACUC approval number: SCXK Hu 2017-0012) which was gifted generously by the National Institute of Parasitic Diseases (NIPD), Chinese Center for Disease Control and Prevention (Shanghai, China). Genomic DNA was extracted from 200 μl diluted blood using the QIAamp DNA Mini Kit (Qiagen, Shanghai, China) according to the manufacturer’s instructions. Sensitivity of fRAA was evaluated on a graded series of
B. microti genomic DNA contents (10
6–10
−1fg/μl). To verify the reproducibility, the sensitivity test was repeated 3 times with the samples collected in 7-day intervals. By fRAA, a perfect detection was obtained with 4 series of diluted samples (10
4, 10
3, 10
2, and 10 fg/μl) of genomic DNA (
Fig. 1C). The lowest detection level was 10 fg/μl genomic DNA. Nested-PCR targeted 18S rRNA gene also detected at least 10 fg/μl in the second PCR (
Fig. 1B). fRAA was less sensitive than the 18S rRNA nested-PCR assay, but had a shorter time to detect samples (20 min vs. more than 2 h).
Other transfusion transmitted parasites elicit similar inflammatory responses as well as clinical manifestations, and they exhibit comparable morphologies under microscope. Sometimes it may be difficult to distinguish these parasites from
B. microti [
17]. We selected 10 apicomplexan parasites to test the specificity of the fRAA assay for
B. microti, including 2 species of Babesia (
B. microti and
B. gibsoni),
Toxoplasma gondii, 2 species of
Leshimania (
L. infantum and
L. donovani),
Entamoeba histolytica, and 4 species of
Plamodium (
P. vivax,
P. falciparum,
P. malariae, and
P. ovale). These parasites were collected or donated from our blood center, and the NIPD, China CDC.
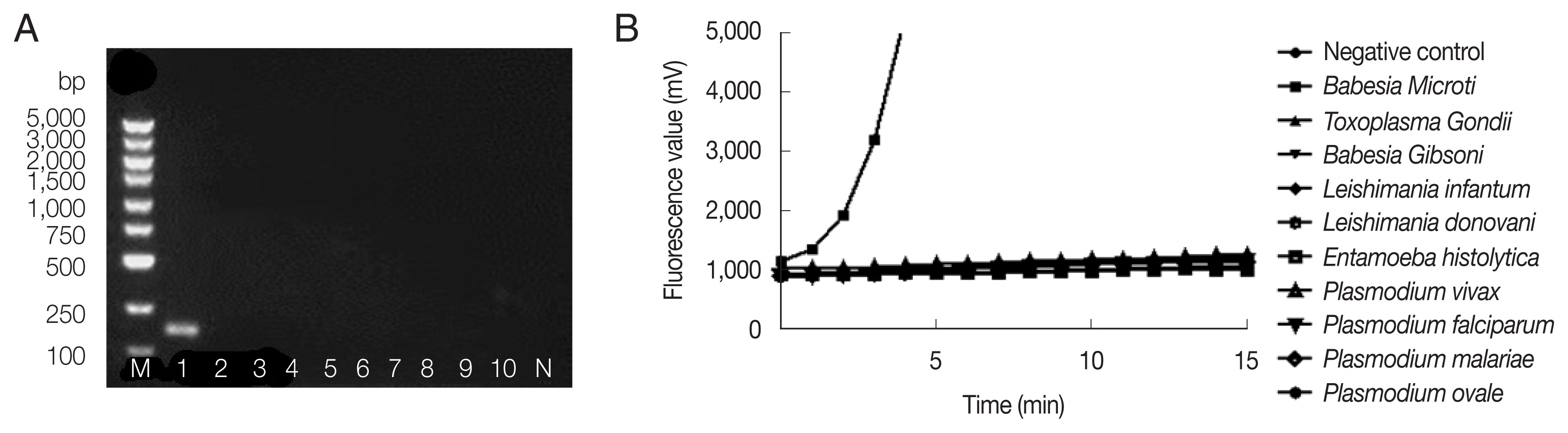
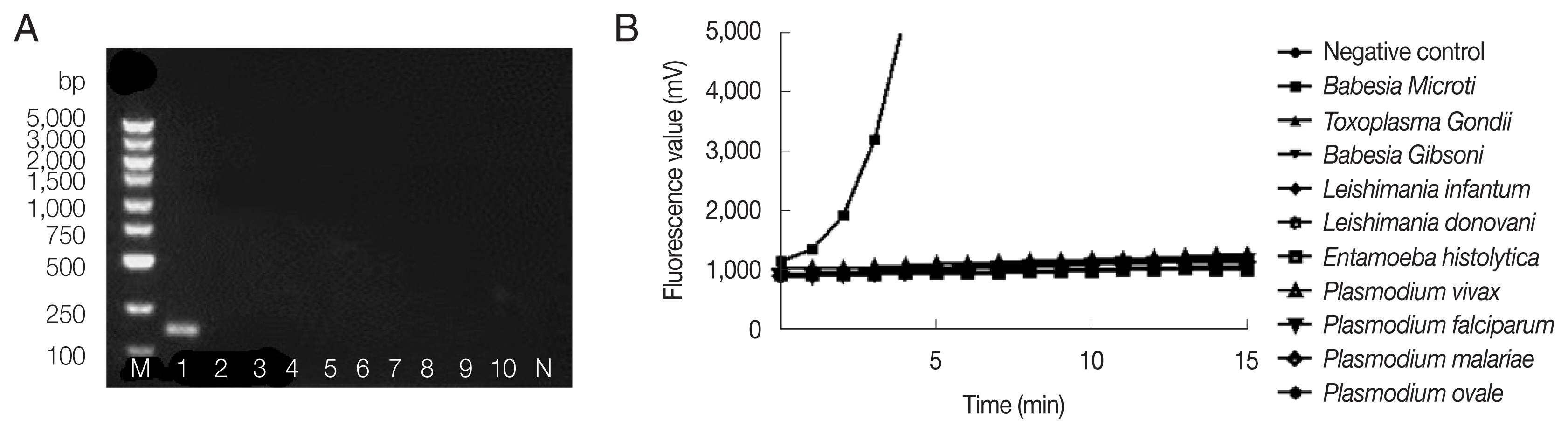
Fig. 2 showed successful amplification of
B. microti genomic DNA, but no bands and fluorescent signals were observed with other pathogenic protozoan genomic DNAs by nested PCR (
Fig. 2A) and fRAA (
Fig. 2B), indicating that fRAA primers and probe were highly specific to
B. microti.
One hundred and sixty blood samples were obtained from our previous work [
18], with the permission of corresponding blood donors and the ethics committee of Jiangsu Province Blood Center (RIB NO. 2019003). These blood donors were all foreign international students who were from East Asia and Africa. These samples, which had been detected negative by nested PCR targeted 18S rRNA gene using different primer pairs [
18], were subjected to fRAA and we found that no sample was positive for
B. microti.
Though the incidence of human babesiosis is very low in China [
19], recent investigations have reported
B. microti infection in small mammals [
20,
21] and humans [
22]. A suspected transfusion transmitted
B. microti infection had also been reported in a transfused patient [
23]. The prevalence of
Babesia spp. among Chinese blood donors was very low [
18,
24], but further investigation and evaluation are needed. This study established a fRAA assay capable of detecting
B. microti within 20 min at 37°C using a portable fluorescence detector. Our fRAA assay exhibited reasonable diagnostic performance and provided a simple, rapid, and reliable method for the detection of
B. microti.
Notes
-
The authors declare that they have no competing interests.
Supplementary Information
ACKNOWLEDGMENTS
This work was supported by Social Development Project (BE2019755) and International Technology Corporation Project (BZ2020003) from the Department of Science and Technology of Jiangsu Province. Supplementary material associated with this article can be found in the online version.
Fig. 1Sensitivity of fRAA assay targeted cox1 compared to nested PCR targeted 18S rRNA gene. (A) Sensitivity evaluated by a recombinant plasmid diluted from 103 to 10 copies/μl. The lowest detection level was 10 copies/μl. (B) Sensitivity of nested PCR as evaluated using the diluted B. microti genomic DNA (Lane1–9: 106, 105, 104, 103, 102, 101, 100, and 10−1 fg/μl and negative control). The second PCR products on the agarose gel showed that the detection sensitivity was 10 fg/μl. (C) Sensitivity of fRAA was evaluated using serially diluted B. microti genomic DNA (104, 103, 102, 101, 100, 10−1 fg/μl and negative control). The sensitivity of fRAA was 10 fg/μl.
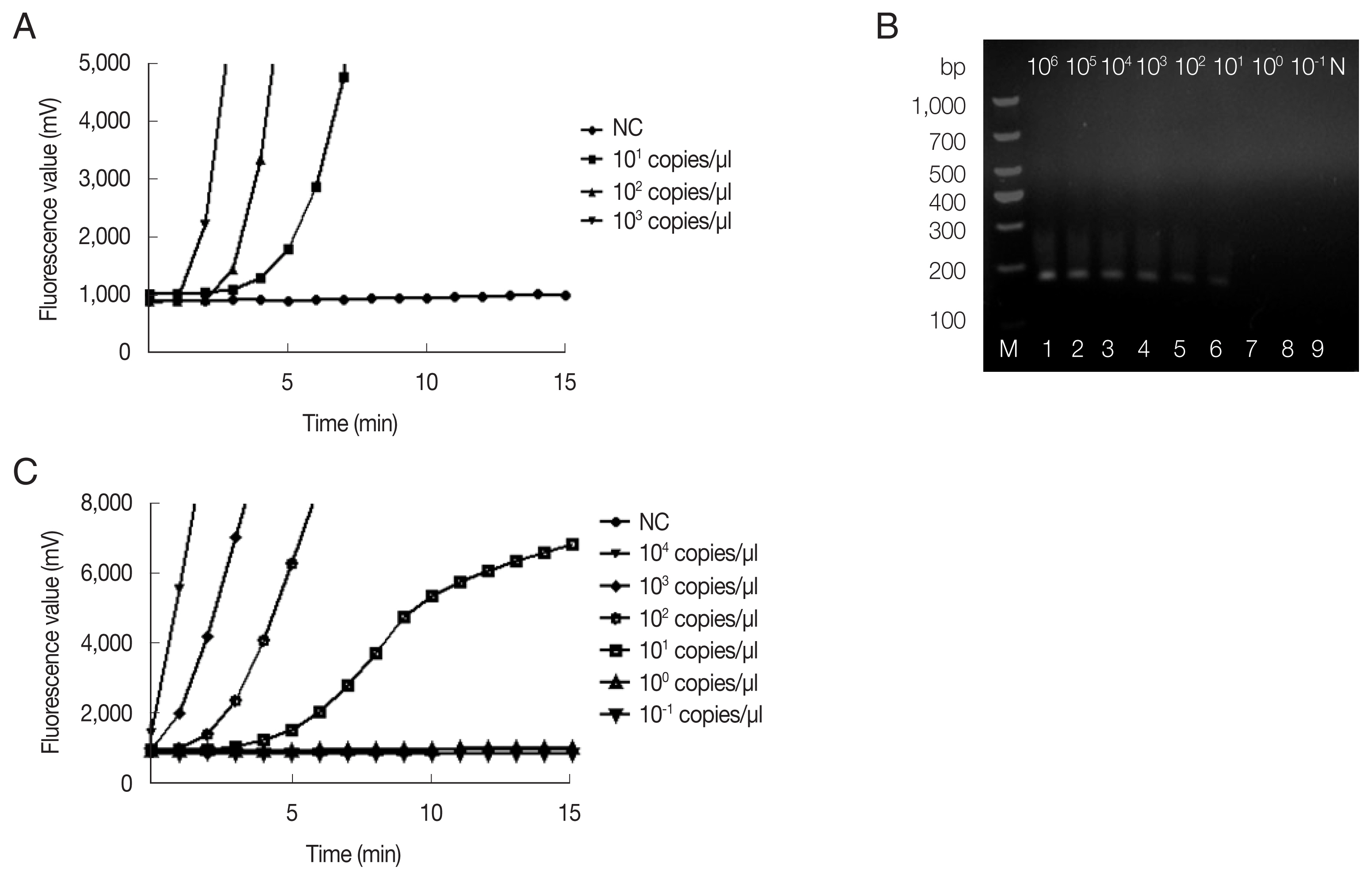
Fig. 2Specificity of nested PCR (A) and fRAA assay (B). (A) Lane 1: B. microti; 2: Toxoplasma gondi; 3: Babesia gibsoni; 4: Leshimania infantum; 5: Leshimania donovani; 6: Entamoeba histolytica; 7: P. vivax; 8: P. malariae; 9: P. falciparum; 10: P. ovale. Only B. microti DNA showed a band on the agarose gel. (B) B. microti DNA showed specific fluorescent signal, while no signal was observed with other parasites DNA.
References
- 1. Madison-Antenucci S, Kramer LD, Gebhardt LL, Kauffman E. Emerging tick-borne diseases. Clin Microbiol Rev 2020;33:e00083-18. https://doi.org/10.1128/CMR.00083-18
- 2. Gumber S, Nascimento FS, Rogers KA, Bishop HS, Rivera HN, Xayavong MV, Devare SG, Schochetman G, Amancha PK, Qvarnstrom Y, Wilkins PP, Villinger F. Experimental transfusion-induced Babesia microti infection: dynamics of parasitemia and immune responses in a rhesus macaque model. Transfusion 2016;56:1508-1519. https://doi.org/10.1111/trf.13521
- 3. Vannier E, Krause PJ. Human babesiosis. N Engl J Med 2012;366:2397-2407. https://doi.org/10.1056/NEJMra1202018
- 4. Xu B, Liu XF, Cai YC, Huang JL, Zhang RX, Chen JH, Cheng XJ, Zhou X, Xu XN, Zhou Y, Zhang T, Chen SB, Li J, Wu QF, Sun CS, Fu YF, Chen JX, Zhou XN, Hu W. Screening for biomarkers reflecting the progression of Babesia microti infection. Parasit Vectors 2018;11:379. https://doi.org/10.1186/s13071-018-2951-0
- 5. Krause PJ, Telford S, Spielman A, Ryan R, Magera J, Rajan TV, Christianson D, Alberghini TV, Bow L, Persing D. Comparison of PCR with blood smear and inoculation of small animals for diagnosis of Babesia microti parasitemia. J Clin Microbiol 1996;34:2791-2794. https://doi.org/10.1128/JCM.34.11.2791-2794.1996
- 6. Levin AE, Williamson PC, Bloch EM, Clifford J, Cyrus S, Shaz BH, Kessler D, Gorlin J, Erwin JL, Krueger NX, Williams GV, Penezina O, Telford SR, Branda JA, Krause PJ, Wormser GP, Schotthoefer AM, Fritsche TR, Busch MP. Serologic screening of United States blood donors for Babesia microti using an investigational enzyme immunoassay. Transfusion 2016;56:1866-1874. https://doi.org/10.1111/trf.13618
- 7. Teal AE, Habura A, Ennis J, Keithly JS, Madison-Antenucci S. A new real-time PCR assay for improved detection of the parasite Babesia microti. J Clin Microbiol 2012;50:903-908. https://doi.org/10.1128/JCM.05848-11
- 8. Stanley J, Stramer SL, Erickson Y, Cruz J, Gorlin J, Janzen M, Rossmann SN, Straus T, Albrecht P, Pate LL, Galel SA. Detection of Babesia RNA and DNA in whole blood samples from US blood donations. Transfusion 2021;6:1317-1325. https://doi.org/10.1111/trf.16617
- 9. Moritz ED, Winton CS, Tonnetti L, Townsend RL, Berardi VP, Hewins ME, Weeks KE, Dodd RY, Stramer SL. Screening for Babesia microti in the U.S. Blood Supply. N Engl J Med 2016;375:2236-2245. https://doi.org/10.1056/NEJMoa1600897
- 10. Goodell AJ, Bloch EM, Krause PJ, Custer B. Costs, consequences, and cost-effectiveness of strategies for Babesia microti donor screening of the US blood supply. Transfusion 2014;7(54):2245-2257. https://doi.org/10.1111/trf.12805
- 11. Wu F, Cai YC, Qin Z, QAL , Lu Y, Chen SH, Wu XP, Chen JX. 2016;Development of loop-mediated isothermal amplification (LAMP) assay combined with FTA card for detecting Babesia microti. Chin J Zoonoses 2016;32:435-441. (in Chinese). https://doi.org/10.3969/j.issn.1002-2694.2016.05.004
- 12. Nie Z, Zhao Y, Shu X, Li D, Ao Y, Li M, Wang S, Cui J, An X, Zhan X, He L, Liu Q, Zhao J. Recombinase polymerase amplification with lateral flow strip for detecting Babesia microti infections. Parasitol Int 2021;83:102351. http://doi.org/10.1016/j.parint.2021.102351
- 13. Hikosaka K, Watanabe Y, Tsuji N, Kita K, Kishine H, Arisue N, Palacpac NM, Kawazu S, Sawai H, Horii T, Igarashi I, Tanabe K. Divergence of the mitochondrial genome structure in the apicomplexan parasites, Babesia and Theileria. Mol Biol Evol 2010;27:1107-1116. http://doi.org/10.1093/molbev/msp320
- 14. Hikosaka K, Tsuji N, Watanabe Y, Kishine H, Horii T, Igarashi I, Kita K, Tanabe K. Novel type of linear mitochondrial genomes with dual flip-flop inversion system in apicomplexan parasites, Babesia microti and Babesia rodhaini. BMC Genomics 2012;13:622. http://doi.org/10.1186/1471-2164-13-622
- 15. Persing DH, Mathiesen D, Marshall WF, Telford SR, Spielman A, Thomford JW, Conrad PA. Detection of Babesia microti by polymerase chain reaction. J Clin Microbiol 1992;30:2097-2103. https://doi.org/10.1128/jcm.30.8.2097-2103.1992
- 16. Kamau E, Tolbert LS, Kortepeter L, Pratt M, Nyakoe N, Muringo L, Ogutu B, Waitumbi JN, Ockenhouse CF. Development of a highly sensitive genus-specific quantitative reverse transcriptase real-time PCR assay for detection and quantitation of Plasmodium by amplifying RNA and DNA of the 18S rRNA genes. J Clin Microbiol 2011;49:2946-2953. https://doi.org/10.1128/JCM.00276-11
- 17. Zhou X, Xia S, Huang JL, Tambo E, Zhuge HX, Zhou XN. Human babesiosis, an emerging tick-borne disease in the People’s Republic of China. Parasit Vectors 2014;7:509. http://doi.org/10.1186/s13071-014-0509-3
- 18. Lin H, Ji YH, Chen XL, Zhu SW, Sun J, Huang CY, Jiao YJ. Sero-prevalence of Babesia infection among blood donors in Jiangsu province. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi 2019;31:516-518. (in Chinese). http://doi.org/10.16250/j.32.1374.2017209
- 19. Chen Z, Li H, Gao X, Bian A, Yan H, Kong D, Liu X. Human babesiosis in China: a systematic review. Parasitol Res 2019;118:1103-1112. http://doi.org/10.1007/s00436-019-06250-9
- 20. Gao ZH, Huang TH, Jiang BG, Jia N, Liu ZX, Shao ZT, Jiang RR, Liu HB, Wei R, Li YQ, Yao HW, von Fricken ME, Jiang JF, Du CH, Cao WC. Wide distribution and genetic diversity of Babesia microti in small mammals from Yunnan province, Southwestern China. PLoS Negl Trop Dis 2017;11:e0005898. http://doi.org/10.1371/journal.pntd.0005898
- 21. Wei CY, Wang XM, Wang ZS, Wang ZH, Guan ZZ, Zhang LH, Dou XF, Wang H. High prevalence of Babesia microti in small mammals in Beijing. Infect Dis Poverty 2020;9:155. http://doi.org/10.1186/s40249-020-00775-3
- 22. Zhou X, Li SG, Wang JZ, Huang JL, Zhou HJ, Chen JH, Zhou XN. Emergence of human babesiosis along the border of China with Myanmar: detection by PCR and confirmation by sequencing. Emerg Microbes Infect 2014;3:e55. http://doi.org/10.1038/emi.2014.55
- 23. Chen Y, Yan D, Zhang YC. Transfusion-associated babesiosis in China: a case report. Transfus Apher Sci 2020;59:102902. http://doi.org/10.1016/j.transci.2020.102902
- 24. Bloch EM, Yang Y, He M, Tonnetti L, Liu Y, Wang J, Guo Y, Li H, Leiby DA, Shan H. A pilot serosurvey of Babesia microti in Chinese blood donors. Vox Sang 2018;113:345-349. http://doi.org/10.1111/vox.12648