Dear Editor,
Multiple infections with small liver flukes and minute intestinal flukes are the serious public health concern in the lower Mekong basin [
1,
2]. Although the epidemiological survey for those trematode infections are primarily carried out based on copro-parasitological examination, detection/identification of fecal eggs/worms is a tedious job and often problematic because of the morphological similarities of eggs/worms. Along with the popularization of PCR-sequencing methods, copro-DNA diagnosis and molecular phylogenetic identification/speciation have been introduced in epidemiological studies. Among various genes and non-coding lesions of nuclear and mitochondrial DNAs, mitochondrial cytochrome c oxidase subunit I (COXI) is one of the most widely used inter- and intra-species marker. Using COXI and some other markers, Lee and his colleagues performed molecular phylogenetic analyses on small liver flukes (Lee SU, Huh S. Variation of nuclear and mitochondrial DNAs in Korean and Chinese isolates of
Clonorchis sinensis. Korean J Parasitol 2004; 42: 145-148) and on minute intestinal flukes (Lee SU, Huh S, Sohn WM, Chai JY. Sequence comparisons of 28S ribosomal DNA and mitochondrial cytochrome c oxidase subunit I of
Metagonimus yokogawai,
M. takahashii and
M. miyatai. Korean J Parasitol 2004; 42: 129-135).
The COX1 gene sequences appeared in those articles are;
Clonorchis sinensis (AF184619, AF181889, AF188122),
Metagonimus yokogawai (AF096230),
Metagonimus takahashii (AF096231),
Metagonimus miyatai (AF096232),
Pygidiopsis summa (AF181884), and
Stellantchasmus falcatus (AF181887). In addition, Park [
3] compared his COXI sequence of Opisthorchis viverrini Laotian isolate (AY055 382) to those of
Gymnophalloides seoi (AF096234) and
Neodiplostomum seoulense (AF096233) registered in the DNA database (Lee et al. unpublished).
For the phylogenetic analyses of COXI gene of minute intestinal flukes of our own data, we have downloaded all those above mentioned COXI of Lee et al. and aligned them including our own COXI sequence of
Haplorchis taichui (EF055885) [
4] and
Paragonimus bangkokensis (AB354227) [
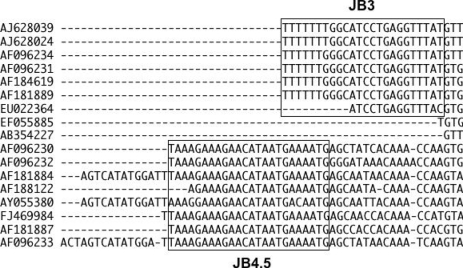
5]. Surprisingly, those sequence data were divided into 2 distinct groups without any similarities (
Fig. 1). Eventually, we realized that this astonishing result is due to the reverse complementary sequences of COXI data deposited by Lee et al. (in the bottom half of the figure). We also noticed similar mixed-up deposition of the forward and reverse sequences of COXI gene of
Fasciola spp., which were also included in
Fig. 1 (AJ628024, AJ628039, FJ469984; Zhu XQ et al. unpublished).
For the determination of partial COX1 sequences of Platyhelminthes, the primer set of JB3 (5'-TTT TTT GGG CAT CCT GAG GTT TAT-3') and JB4.5 (5'-TAA AGA AAG AAC ATA ATG AAA ATG-3') [
6] was widely used for investigating the inter- and intra-species variations of trematodes and cestodes. We noticed the mixed-up of the forward and reverse COXI sequences by Lee et al. as well as Zhu et al. because of the presence of the characteristic feature of this primer set (boxed in
Fig. 1) in the sequences. The primer sequence should be deleted from the sequence data because it is not always identical with the real DNA sequence of the gene and the inclusion of the primer sequences sometimes causes the misreading in phylogenetic analyses [
7]. In 3 reverse sequences, AF181884, AY055380, and AF096233 seems to contain also the partial sequence of the cloning vector, which should be trimmed off before deposition.
In general, raw data of forward and reverse sequences obtained from the sequencer should be aligned manually by cross-checking of the wave patterns because some 10-20 bases downstream from the forward primer and upstream from the reverse primer often contain erroneous base pairs [
8]. Deposition of the reverse sequence means that those sequences were not aligned against forward sequence and not quite reliable.
Since each sequence data in GenBank are opened for the public use, an accuracy of the sequence data is critically important for the mutual reliability of the scientists. The scientists should aware how to deposit accurate sequence data to the DNA data base. The reappraisal and correction of those sequences mentioned above is urgently necessary.
References
- 1. Radomyos B, Wongsaroj T, Wilairatana P, Radomyos P, Praevanich R, Meesomboon V, Jongsuksuntikul P. Opisthorchiasis and intestinal fluke infections in northern Thailand. Southeast Asian J Trop Med Public Health 1998;29:123-127.
- 2. Chai JY, Han ET, Shin EH, Sohn WM, Yong TS, Eom KS, Min DY, Um JY, Park MS, Hoang EH, Phommasack B, Insisiengmay B, Lee SH, Rim HJ. High prevalence of Haplorchis taichui, Phaneropsolus molenkampi, and other helminth infections among people in Khammouane Province, Lao PDR. Korean J Parasitol 2009;47:243-247.
- 3. Park GM. Genetic comparison of liver flukes, Clonorchis sinensis and Opisthorchis viverrini, based on rDNA and mtDNA gene sequences. Parasitol Res 2007;100:351-357.
- 4. Thaenkham U, Visetsuk K, Dung DT, Waikagul J. Discrimination of Opisthorchis viverrini from Haplorchis taichui using COI sequence marker. Acta Trop 2007;103:26-32.
- 5. Doanh PN, Shinohara A, Horii Y, Yahiro S, Habe S, Vannavong N, Strobel M, Nakamura S, Nawa Y. Morphological differences and molecular similarities between Paragonimus bangkokensis and P. harinasutai. Parasitol Res 2009;105:429-439.
- 6. Bowles J, Hope M, Tiu WU, Liu SX, McManus DP. Nuclear and mitochondrial genetic markers highly conserved between Chinese and Philippine Schistosoma japonicum. Acta Trop 1993;55:217-229.
- 7. Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Env Microbiol 2008;74:2461-2470.
- 8. Automated DNA Sequencing: Chemistry Guide. 7. Data Evaluation and Troubleshooting. 2000, Foster, California, USA. Applied Biosystems.
Fig. 1The DNA sequence alignment of the partial COXI gene of some trematodes obtained from the GenBank. Seven sequences from the top are the forward strands with JB3 primer sequence, and 2 in the middle are the forward strands without primer. Eight sequences in the bottom are the reverse strands with JB4.5 primer sequence, and 3 of them had additional sequence of the cloning vector. The JB3 and JB4.5 primer portion were indicated by square frames.
Citations
Citations to this article as recorded by