Abstract
The identification, characterization and quantification of Plasmodium sp. genetic polymorphism are becoming increasingly important in the vaccine development. We investigated polymorphism of Plasmodium vivax GAM-1 (PvGAM-1) gene in 30 Korean isolates. The polymorphic region of the PvGAM-1 gene, corresponding to nt 3792-4029, was amplified using polymerase chain reaction (PCR) followed by sequencing. All of the P. vivax Korean isolates were one type of GAM-1 gene, which were identical to that of the Belem strain. It is suggested that PvGAM-1 could not be used as a genetic marker for identifying or classifying P. vivax Korean isolates. It revealed that the polymorphic pattern was acquired basically by duplication and modification or deletion event of a 33 bp-motif fragment ended by poly guanine (G) and that there were at least three complete and one partial 33 bp-motif sequences within the polymorphic region in the longest cases such as those of South Korean and Belem isolates. In addition, we clustered P. vivax isolates with parsimonious criteria on the basis of PvGAM-1 polymorphic patterns (insertion/deletion patterns).
-
Key words: Plasmodium vivax, GAM-1, genotype, polymorphism, Korean isolates
INTRODUCTION
Among four known
Plasmodium species that cause malaria in humans,
P. vivax malaria are not only the most prevalent but also Important to human health. Although
P. vivax is not frequently fatal to human, it is an exhausting, debilitating disease that impairs quality of life and economic productivity. The enormous toll of mortality caused by
P. falciparum has tended to overshadow the public health importance of
P. vivax. For that reason and technical difficulties, relatively little investment has been made in attempts to develop a vaccine against
P. vivax. (
Snewin et al., 1991).
Although large amounts of investment have been made in attempts to develop a vaccine against to
P. falciparum, so far no available form has been developed. One of major problems in vaccine development is antigenic diversity of the vaccine candidate proteins. The critical problem is emerging from the fact that host response to one allele is much less effective against parasites expressing different allelic forms (
Crewther et al., 1996;
Renia et al., 1997). The polymorphism of an immunodominant antigen, a potential malaria vaccine, is rather greater for
P. vivax than for
P. falciparum (
Arnot et al., 1990). Therefore, the polymorphism of a candidate vaccine protein may be serious in
P. vivax rather than
P. falciparum in development of malaria vaccine. The polymorphism study is not only important to establish the antigenic repertoire of isolates from malaria endemic regions but also to elucidate the mechanisms by which antigenic diversity may be generated.
P. vivax malaria had re-emerged after being absent for more than 10 years in the Republic of Korea (
Kho et al., 1999a;
Chai, 1999). In the previous study,
P. vivax Duffy binding protein (PvDBP) and circumsporozoite protein (PvCSP) showed genotypes with at least two new phenotypes among the Korean isolates (
Kho et al., 1999b;
Kho et al., 2001). However, the accurate extent of genetic diversity of
P. vivax Korean isolates is not known at present. This is due to the availability of very few polymorphic markers for studying
P. vivax. The
P. vivax GAM-1 (PvGAM-1), a transmission blocking candidate antigen, was suggested a new polymorphic marker for use in field studies of the
P. vivax parasite population (
Snewin et al., 1995). However, the only available information has been made from the Sri Lankan isolates of
P. vivax.
To add useful information for genetic diversity and rational design of vaccine, polymorphism of PvGAM-1 in the Korean isolates were investigated in this study. Besides, we clustered P. vivax isolates on the basis of insertion/deletion patterns of their PvGAM-1 genes using a phylogenetic approach and suggested a hypothetical evolutionary pathway for Sri Lankan PvGAM-1 polymorphism.
MATERIALS AND METHODS
Isolation of genomic DNA
In 1998, blood samples were collected from 30 P. vivax patients who were detected in Yonchon-gun, Kyonggi-do. All patients were diagnosed by microscopic examination at the Institute of Malariology, Inje University.
P. vivax DNA was extracted from 0.1 ml of EDTA-treated blood samples using QIAamp DNA blood kit (Qlagen, Valencia, CA, USA). The isolated DNA was dissolved in TE buffer (10 mM Tris-HCl, pH 7.4 and 1 mM EDTA, pH 8.0) and stored at -70℃ until use.
Semi-nested polymerase chain reaction (PCR)
Amplification of PvGAM-1 gene by the semi-nested polymerase chain reaction (PCR) was performed in 20 µ1 reaction mixture containing the P. vivax DNA, 200 µM dNTP each, 0.5 pM primer each, 10 mM Tris-HCl, pH 8.0, 50 mM KC1, 1.0 mM MgCl2, and 0.25 units of Taq polymerase (Takara, Kyoto, Japan). The PCR primers were PG5: 5'-CTTCCGCAGGTAC CTCCAAA-3' (nucleotides 3715 to 3734, positions after GenBank accession number X84734 of the Belem sequence) and PGR1: 5'-AGGCATC GAATGACCTGTC -3' (positions 4190 - 4172). We amplified this region by semi-nested PCR using oligonucleotide primers PGF1 (5' -GTGAATGCAGTTCTGTGCG-3' , positions 3737 - 3755 of GenBank No. X84734) and PGR1 after initial PCR. The thermal cycler profile contained 35 cycles of 94, 50 and 72℃ for 1, 1 and 2 min, respectively.
Sequencing and gene analysis
DNA fragments amplified by PCR were separated on 1.2% agarose gel. The amplified DNA was purified from agarose gel using a QIAEX II gel extraction kit (Qiagen, Valencia) following the manufacturer's instruction. The nucleotide sequence was determined by a dideoxynucleotide chain termination method using a sequenase kit (ABI PRISM Dye Terminator Cycle Sequencing Core Kit, Perkin Elmer) and an automated DNA sequencer (Applied Biosystems model 377A, Perkin Elmer). Primers for sequencing were PGF1 and PGR I. Verification of nucleotide substitutions was performed by direct sequencing of newly amplified PCR products from the same isolates.
Preliminary pairwise sequence alignment and comparison were performed using GeneJockey II (Biosoft Co.) and the BLAST program of the NCBI databases (NIH, Bethesda, MD, USA). Multiple sequence alignment was constructed with Clustal-X (
Thomson et al., 1997). WDNASIS (Hitachi, Ver. 2.5, Japan) was used for translating DNA sequences into amino acid sequences and for predicting protein secondary structure. Phylogenetic analysis was conducted with maximum parsimony method of PAUP
* 4.0 (
Swofford, 2001). DNA sequences were used as input data in the phylogenetic analysis and alignment gaps were treated as fifth characters. Sequence data of the Korean isolates were compared with published sequence of
P. vivax including 6 Sri Lankan isolates, Belem strain, and Chesson strain (
Snewin et al., 1995). The PvGAM-1 from Belem strain, was used as an outgroup.
RESULTS
PCR amplifications of PvGAM-1 genes from Korean isolates
According to the blood smear analysis, the parasitemias ranged between 0.004% and 0.28% with mean value of 0.073%, between 208 and 13,888 parasites per reaction. Although in varying concentration, the PvGAM-1 gene was successfully amplified in all cases after two rounds of semi-nested PCR.
A single band of about 450 bp was seen after two rounds of amplification, corresponding to 3737-4190 positions of the Belem strain sequence. Niether size polymorphism nor multiple bands were observed. No specific PCR product was found from uninfected donor blood used as a negative control (data not shown).
Nucleotide sequence analysis on PvGAM-1 polymorphic regions in Korean isolates
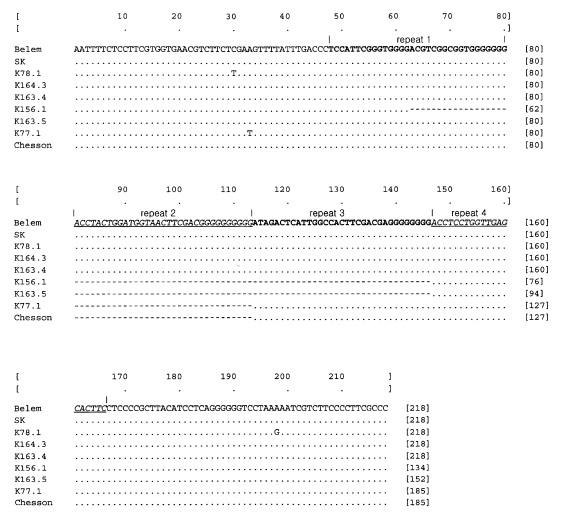
DNA sequences of polymorphic region, corresponding to 3792-4029 positions of the Belem strain sequence, were obtained from thirty Korean isolates. Only one genotype was identified from the thirty isolates and thus any polymorphic phenomena were not found. The resultant sequences were compared to those of the Belem strain, Chesson strain, and Sri Lankan isolates, which showed that PvGAM-1 of Korean isolates was identical to that of the Belem strain (
Fig. 1). The nucleotide sequence of PvGAM-1 from Korean isolate was enrolled in the GenBank Database (accession no. AY050644).
Nine genotypes of PvGAM-1 including 1 South Korean genotype, 6 Sri Lankan isolates, Belem strain, and Chesson strain genotypes reported by Snewin et al. (
1995) were aligned. Through careful observation of the multiple alignment of the PvGAM-1 polymorphic region, we found that the polymorphism (insertion/deletion) was obtained basically by duplication and modification or deletion event of a 33 bp-motif fragment ended by poly guanine (G) residues. The three complete and one partial 33 bp-motif sequences were repeated within polymorphic regions in the longest cases such as those of South Korean and Belem isolates. Sequence similarity among the repeatitive 33 bp-motif sequences within PvGAM-1 polymorphic region of a strain or a isolate were significantly high (
Fig. 2).
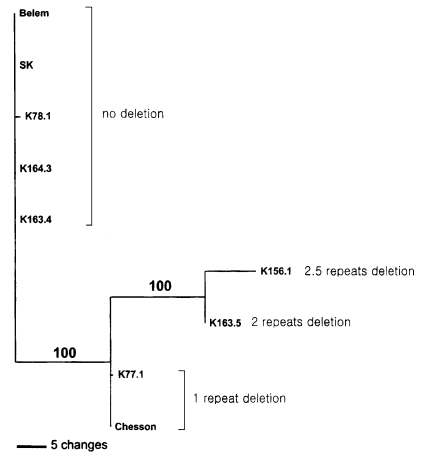
Phylogenetic analysis using maximum parsimony method showed that the examined isolates were clustered along the patterns of insertion/deletion of the motif sequences as shown in
Fig. 3: (Belem, SK, K781, K164.3, K164.4, (K77.1, Chessen, (K156.1, K163.5))).
DISCUSSION
The nested PCR is a powerful technique that detects possibly a low level parasitemias and a ratio of 1:1000 mixed infection (
Contamin et al., 1995). Therefore, the use of semi-nested PCR can result in the ultimate sensitivity. Thirty Korean isolates blood samples used for sequence analysis had no evidence of mixed infection such as multiple band. A high transmission intensity should allow, not only multiclonal infection of parasites, but also new genotypes resulting from frequent out-crossing within mosquito (
Babiker and Walliker, 1997). Therefore, single infection in a patient and one genotype among Korean isolates may imply that malaria transmission intensity of South Korea is very low.
The nucleotide sequences of PvGAM-1 genes from the examined Korean isolates were identical to those of Belem strain (
Fig. 1). This result suggests that the variation of PvGAM-1 may occur very seldom. It was proved that the polymorphic region of PvGAM-1 is not the epitope, which is involved in transmission blocking immunity (
Udagama et al., 1987). A polymorphic region is flanked by conserved sequences. This phenomenon also showed in the PvDBP gene, which is the parasite ligand for Duffy-glycoprotein receptor in human erythrocytes. It was suggested that the variation of the polymorphic region flanked by a central conserved region may be related with the immune pressure and selection (
Tsuboi et al., 1994;
Kho et al., 2001). According to the hypothesis, the region of the Korean isolate is less likely to be subjected to immune pressure or selection. Immune pressure and selection may be closely related to intensity of malaria transmission. Therefore the difference in the number of PvGAM-1 variants between the South Korean and the Sri Lankan isolates may be to the difference in the intensity of malaria transmission. Although the Intensity of malaria transmission in the Republic of Korea has not been scientifically measured, it is believed that the opportunity of malaria being transmitted by mosquitoes might be strictly limited since the transmission is possible only during the limited season from May to October and the transmission ability of
Anopheles vector mosquito may be extremely low (
Ree, 2000;
Ree et al., 2001). It is likely that genetic change of polymorphic region may occur more slowly in the Republic of Korea than in the high-endemic areas because of the epidemic characteristics of malaria.
In a number of cases, rapid evolving DNA regions have often given us critical clues to understand gene evolution mechanism (
Choe et al., 1999a,
1999b;
Ryu et al., 1999;
Hwang et al., 2000). This result showed that the different number of 33 bp-motif sequences causes PvGAM-1 gene polymorphism of Sri Lankan
P. vivax isolates. The 33 bp-motif was integrated between G and AC nucleotides of aspartic acid codon (GAC), which did not interrupt the open reading frame of PvGAM-1 gene. The motif always started with AC or AU and ended with G. Thus, the joint regions of both ends form always aspartic acid codon, GAC or GAU. Gene size exapansion or reduction by insertion/deletion event of a specific motif has been reported in many cases (
Choe et al., 1999a,
1999b;
Ryu et al., 1999). It is known that such a evolutionary pattern mainly appears in fast evolving regions within a gene.
According to the result of protein secondary structure prediction of the polymorphic region, almost all of this region did not form any sheet or helix structure but were related only with turning of the polypeptide chain (data not shown).
The present finding suggests that the variation of PvGAM-1 may occur very seldom In the Republic of Korea, thus PvGAM-1 may not be a good polymorphic genetic marker in Korean P. vivax isolates. Through further studies, if additional PvGAM-1 gene sequences from P. vivax isolates over a worldwide range become known and their polymorphic regions are carefully compared, we could have a general speculation related with its utility as a genetic marker and its evolutionary mechanism.
Notes
-
This study was supported by Korea Research Foundation made in the program year of 1998.
References
- 1. Arnot DE, Stewart MJ, Barnwell JW. Antigenic diversity in Thai Plasmodium vivax circumsporozoite proteins. Mol Biochem Parasitol 1990;43:147-149.
- 2. Babiker HA, Walliker D. Current views on the population structure of Plasmodium falciparum: Implication for control. Parasitol Today 1997;13:262-267.
- 3. Bolad A, Berzins K. Antigenic diversity of Plasmodium falciparum and antibody-mediated parasite neutralization. Scand J Immunol 2000;52:233-239.
- 4. Chai JY. Re-emerging Plasmodium vivax malaria in the Republic of Korea. Korean J Parasitol 1999;37:129-143.
- 5. Choe CP, Hancock JM, Hwang UW, Kim W. Analysis of the primary sequence and secondary structure of the unusually long SSU rRNA of the soil bug, Armadillidium vulgare. J Mol Evol 1999a;49:798-805.
- 6. Choe CP, Hwang UW, Kim W. Putative secondary structure of unusually long strepsipteran SSU rRNAs and its phylogenetic Implications. Mol Cells 1999b;9:191-199.
- 7. Contamin H, Fandeur T, Bonnefoy S, Skouri F, Ntoumi F, Mercereau-Puijalon O. PCR typing of field isolates of Plasmodium falciparum. J Clin Microbiol 1995;33:944-951.
- 8. Crewther PE, Matthew ML, Flegg RH, Anders RF. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infect Immun 1996;64:3310-3317.
- 9. Hwang UW, Ree HI, Kim W. Evolution of hypervariable regions, V4 and V7, of insect 18S rRNA and their phylogenetic implications. Zool Sci 2000;17:111-121.
- 10. Kho WG, Jang JY, Hong ST, Lee HW, Lee WJ, Lee JS. Border malaria characters of reemerging vivax malaria in the Republic of Korea. Korean J Parasitol 1999a;37:71-76.
- 11. Kho WG, Park YH, Chung JY, et al. Two new genotypes of Plasmodium vivax circumsporozoite protein found in the Republic of Korea. Korean J Parasitol 1999b;37:265-270.
- 12. Kho WG, Chung JY, Sim EJ, Kim DW, Chung WC. Analysis of polymorphic regions of Plasmodium vivax Duffy binding protein of Korean isolates. Korean J Parasitol 2001;39:143-150.
- 13. Ree HI. Unstable vivax malaria in Korea. Korean J Parasitol 2000;38:119-138.
- 14. Ree HI, Hwang UW, Lee IY, Kim TE. Daily survival and human blood Index of Anopheles sinensis, the vector species of malaria in Korea. J Am Mosq Control Assoc 2001;17:67-72.
- 15. Renia L, Ling IT, Marussig M, Miltgen F, Holder AA, Mazier D. Immunization with a recombinant C-terminal fragment of Plasmodium yoelii merozoite surface protein 1 protects mice against homoloous but not heterologous P. yoelii sporozoite challenge. Infect Immun 1997;65:4419-4423.
- 16. Ryu SH, Do YK, Hwang UW, Choe CP., Kim W. Ribosomal DNA intergenic spacer of the swimming crab, Charybdis Japonica. J Mol Evol 1999;49:806-809.
- 17. Snewin VA, Longacre S, David PH. Plasmodium vivax: older and wiser? Res Immunol 1991;142:631-636.
- 18. Snewin VA, Khouri E, Wattavidanage J, et al. A new polymorphic marker for PCR typing of Plasmodium vivax parasites. Mol Biochem Parasitol 1995;71:135-138.
- 19. Swofford DL. PAUP*4.0. Phylogenetic analysis using parsimony (* and other methods). 2001, Version 4. Sunderland, MA. Sinauer Associates.
- 20. Tsuboi T, Kappe SH, al-Yaman F, Adams JH. Natural variation within the principal adhesion domain of the Plasmodium vivax duffy binding protein. Infect Immun 1994;62:5581-5586.
- 21. Thomson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL-X-windows interface: Flexible starategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acid Res 1997;25:4876-4882.
- 22. Udagama PV, David PH, Peiris JS, Ariyaratne YG, Perera KL, Mendis KN. Demonstration of antigenic polymorphism in Plasmodium vivax malaria with a panel of 30 monoclonal antibodies. Infect Immun 1987;55:2604-2611.
Fig. 1Nucleotide sequence of Korean isolates (SK) in the polymorphic region (corresponding to nt 3792 - 4029) of the PvGAM-1 gene, comparing with Belem strain, Chesson strain, and Sri Lankan isolates (K78.1, C164.3, C163.4, K156.1, C163.5. and K77.1) (
Snewin et al., 1995). Gaps indicated by dashes (- - -) are introduced into the sequence to optimize the alignment. Dot marks the same nucleotide sequences with those of the first line, Belem strain. Bold and underlined italic type letters are repeat units (basically 33 bp), which consist of repeat 1, repeat 2, repeat 3 and repeat 4 in order. Except fourth truncated repeat (20 bp), the others are 33 bp in length.

Fig. 2Alignment of four repeat (motif) sequences observed In the longest polymorphic regions of PvGAM-1 gene such as those from Belem strain or South Korean isolates. Fourth motif is truncated and a dash (-) in the truncated region marks an alignment gap. Dots mark the same nucleotide sequences with those of the first line (consensus sequences). Nucleotide sequences of all the repeats are highly conserved.

Fig. 3Maximum parsimonious tree reconstructed with nucleotide sequences of polymorphic regions of PvGAM-1 genes from South Korean isolates (SK), Belem strain, Chesson strain, and Sri Lankan isolates (K78.1, C164.3, C163.4, K156.1, C163.5, and K77.1). Alignment gaps were treated as fifth characters. Numbers of above branches indicate bootstrapping values obtained with 1000 replicates (tree length = 87. CI = 1.0). Of 218 total characters. 131 and 66 characters are constant and parsimoniously informative, respectively.