Abstract
The onset, severity, and ultimate outcome of malaria infection are influenced by parasite-expressed virulence factors and individual host responses to these determinants. In both humans and mice, liver injury is involved after parasite entry, which persists until the erythrocyte stage after infection with the fatal strain Plasmodium falciparum (Pf). Hepatocyte growth factor (HGF) has strong anti-apoptotic effects in various kinds of cells, and also has diverse metabolic functions. In this work, Pf-subtilisin-like protease 2 (Pf-Sub2) 5'untranslated region (UTR) was analyzed and its transcriptional activity was estimated by luciferase expression. Fourteen TATA boxes were observed but only one Oct-1 and c-Myb were done. In addition, host HGF interaction with Pf-Sub2 was evaluated by co-transfection of HGF- and Pf-Sub2-cloned vector. Interestingly, -1,422/+12 UTR exhibited the strongest luciferase activity but -329 to +12 UTR did not exhibit luciferase activity. Moreover, as compared with the control of unexpressed HGF, the HGF protein suppressed luciferase expression driven by the 5'untranslated region of the Pf-Sub2 promoter. Taken together, it is suggested that HGF controls and interacts with the promoter region of the Pf-Sub2 gene.
-
Key words: Plasmodium falciparum, malaria, subtilisin-like protease 2, hepatocyte growth factor, 5'untranslated reagion, promoter
INTRODUCTION
Malaria, which is caused by infection with the hemoprotozoan parasite
Plasmodium, afflicts approximately 500 million people each year, leading to 2.7 million deaths annually worldwide [
1]. Malaria results in diverse clinical outcomes presumably because of dynamic relationships between parasite-expressed virulence factors and individual host responses to these determinants [
2]. The last few years have seen a rapid growth in the number of reported genetic associations with susceptibility and resistance to malaria in the host; they are mainly variants in erythrocyte, cytoadherence factor, immune system, and inflammatory genes [
3,
5]. However, molecular basis of malaria infection remains incompletely defined. Identification of genetic factors in both the host and the parasite involved in malaria susceptibility and resistance would lead not only to a greater understanding of this complex disease, but potentially also to the development of effective medical interventions [
2,
6].
Interactions between
Plasmodium and its host hepatocyte factors are of great research interest [
7-
9]. On one hand, it is well established that malaria sporozoites exploit hepatocyte growth factor (HGF) and its receptor MET signaling during host invasion [
10]. On the other hand,
Plasmodium is a life cycle-specific disease that includes liver injury at the erythrocyte stage of the parasite [
11]. Indeed, both humans and mice suffer from liver injury after infection with the fatal strains
Plasmodium falciparum (
Pf) and
P. berghei (
Pb) [
12,
13]. Because
Pb merozoites do not parasitize liver parenchymal cells in mice, it was proposed that the mechanism of
Pb-induced liver injury is likely to be caused by the local production of cytokines that activate lymphocytes residing in the liver [
14]. Pathologically, malaria-induced mouse liver injury is characterized by dense infiltration of lymphocytes, and the presence of apoptotic and necrotic hepatocytes [
15].
Plasmodium subtilisin-like protease 2 (subtilase, Sub2) is an essential integral membrane serine protease in both
Pf and
Pb. It functions in the shedding of the ectodomain components of 2 surface proteins, membrane surface protein 1 (MSP1), and apical membrane antigen 1 (AMA1), upon host invasion [
16-
18]. The importance of the Sub2 gene is evidenced by the demonstration that gene-knock out parasites cannot be recovered [
19].
The correlation of HGF and malaria infection is not well understood. In this study, to understand the relationship between the malaria infection-related protein Pf-Sub2 and HGF, the Pf-Sub2 5'untranslated region (UTR) was analyzed and its promoter activity was estimated. In addition, the influence of HGF on its promoter activity was analyzed.
MATERIALS AND METHODS
Parasite manipulations
Pf 3D7 strain was obtained from Malaria Research and Reference Reagent Resource Center (MR4; Manassas, Virginia, USA;
http://www.mr4.org). The
Pf 3D7 culture condition was set according to the MR4 standard protocol. Parasitemia was calculated by counting at least 1,000 cells following staining with Diff Quick (Sysmex, Kobe, Japan).
Cell culture
Cell culture media and components, including fetal bovine serum (FBS), were obtained from Invitrogen (Carlsbad, California, USA). All cells were purchased from the American Type Culture Collection (ATCC, Rockville, Maryland, USA) and were routinely cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% FBS. HeLa, Huh-7, HepG2, and 293T cells were used for the experiments.
DNA isolation and plasmid vector construction
All plasmids were purified using JETstar2.0 mini and maxiprep Kit (GenoMed, St. Louis, Missouri, USA) according to the manufacturer's instructions. Pf 3D7 small scale gDNA isolation followed the MR4 protocol. A construct for luciferase activity was based on a pGL3-Basic vector (Promega, Madison, Wisconsin, USA). Two different 5'upstream sequences flanking the Pf-Sub2 gene were amplified from Pf 3D7 gDNA with primers. The primers used for -2,567/+32 UTR were F (5'-ATAGGTACCTAGAATAGAATAAAATAAATAACC-3'; KpnI site underlined) and R (5'-ATAAAGCTTATCAAGGAAACCACATAA-3'; HindIII site underlined). The primers used for -1,422/+12 UTR were F (5'-AAGGTACCATGGTATGAGTTCTTTATA-3'; KpnI site underlined) and R (5'-ATGCTAGCAATATTCAGCATTATACGGA-3'; NheI site underlined). PCR was performed with a TGRADIEN thermocycler (Biometra, Goettingen, Germany) and the amplified UTR sequences were cloned into a pMDT vector (Takara Bio, Shiga, Japan) and subcloned into a pGL3-Basic vector with KpnI and NheI/HindIII sticky end ligation, generating pGL3-Basic-Pf-Sub2 5'UTR. The other upstream inserts (-329/+12, -668/+12 and -739/+12) were cloned into a pGL3-Basic vector by blunt restriction sites (EcoRV, SnaBI and SwaI) located in the 1.5 kb region upstream of the luciferase gene. The internal control was pSV-β-galactosidase (Promega). All clones were analyzed by restriction mapping and were sequenced by Macrogen (Seoul, South Korea). To study the interactions between Pf-Sub2 5'UTR and HGF, the HGF gene was cloned into pcDNA3 Flag HA vector using the enzymes of BamHI and XhoI.
Transfection and luciferase assay
pGL3-Basic-Pf-Sub2 5'UTR and pcDNA3 Flag HA HGF vector were co-transfected at a ratio of 2:1 by a standard calcium phosphate co-precipitation method in 6-well plates. Each sample was analyzed in duplicate and each experiment was repeated twice. Luciferase activity was measured using a model LB96V microplate luminometer (Berthold Technologies, Bad Wildbad, Germany) according to the manufacturer's instructions. Luciferase activity was normalized with respect to β-galactosidase activity to correct deviation due to transfection efficiencies and cell numbers.
SDS-PAGE and western blot analysis
Cells were harvested and lysed with RIPA buffer (Pierce, Rockford, Illinois, USA) with freshly added protease inhibitor cocktail (Sigma-Aldrich, St. Louis, Missouri, USA). One hundred nanograms of cell lysate were diluted with a loading buffer that contained a reducing agent, and the proteins were separated by 12% SDS-PAGE and transferred to a polyvinylidene fluoride membrane. Immunoblotting was performed with a 1:200 dilution in 3% bovine serum albumin (BSA) of rabbit polyclonal anti-HA (Dako). The secondary antibody was a 1:1,000 dilution in PBS of polyclonal goat anti-rabbit immunoglobulin G (IgG) conjugated with horseradish peroxidase (HRP) (Dako, Glostrup, Denmark). For proliferating cell nuclear antigen (PCNA) examination, anti-PCNA/HRP monoclonal antibody was used (Dako).
RESULTS
Analysis of Pf-Sub2 promoter regions
Transcription elements in
Pf-Sub2 5'UTR were analyzed. Several transcriptional elements were identified within about 1,453 bp and are listed in
Table 1. Most transcription elements were 14 TATA boxes but only 1 Oct-1 and c-Myb were observed.
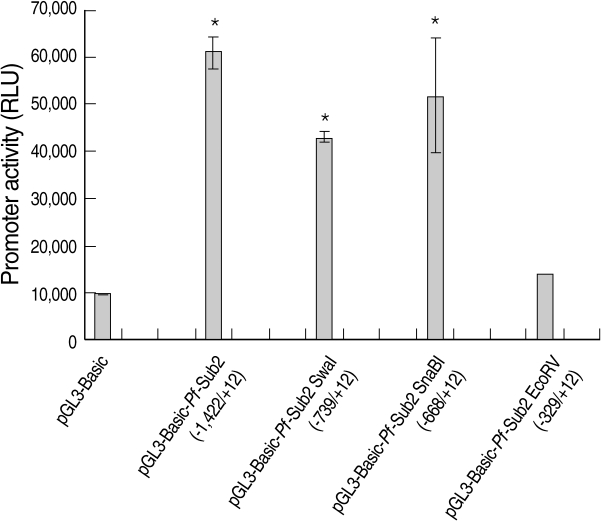
We assessed the
Pf-Sub2 upstream -1,422/+12 UTR 5'promoter region from -1,422, -739, -668 and -329 to +12. The pGL3-Basic vector cloned with
Pf-Sub2 5'UTR driving the luciferase gene and pGL3-Basic-
Pf-Sub2 5'UTR was transfected into 293T cells, and its luciferase activities were measured (
Fig. 1). In 293T cells, luciferase activities in -1,422/+12, -739/+12 and -668/+12 5'UTR were 4-7-fold higher than in the pGL3-Basic vector (
P < 0.05). Among them, -1,422/+12 UTR exhibited the strongest luciferase activity but -329 to +12 UTR did not exhibit luciferase activity. Luciferase expression was not derived from transfection efficiency. In fact, transfection efficiency of the pGL3-Basic vector determined by use of the SV40 promoter was 1 × 10
7 in 293T cells.
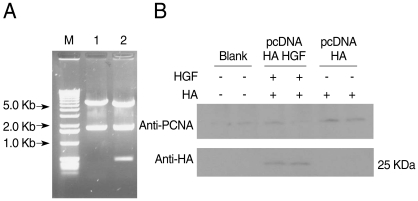
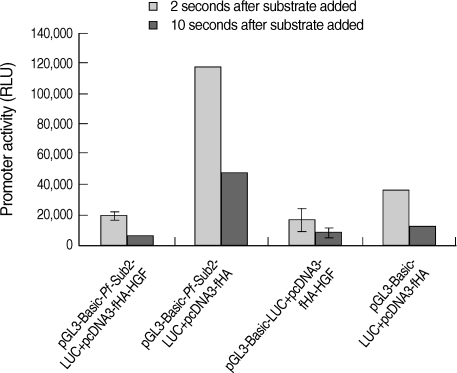
HGF genes were cloned into pcDNA3 HA vector and confirmed by restriction enzyme digestion (
Fig. 2A). Western blotting was performed to analyze whether pcDNA3 Flag HA HGF vector was expressed in 293T cells after it was transfected using FuGENE 6 (
Fig. 2B). HGF expression was observed, with about a 25 kDa band. To analyze the influence of HGF on the transcriptional activity of 5'UTR of
Pf-Sub2 in vitro, co-transfection of pGL3-Basic-
Pf-Sub2 5'UTR (-1,422/+12) and pcDNA3 Flag HA HGF vector was performed in 293T cells. The influence of HGF protein onto
Pf-Sub2 5'UTR (-1,422/+12) was estimated by the expression of a luciferase gene directed by the
Pf-Sub2 5'UTR (
Fig. 3). As compared with the control of unexpressed HGF, the HGF protein suppressed luciferase expression by a factor of about 6. It is suggested that HGF controls and interacts with the promoter region of
Pf-Sub2 gene.
DISCUSSION
Plasmodium infects liver tissue, so to understand malaria infection it is necessary to understand the relation between hepatocytes and malaria. However, this process is poorly understood. Sporozoites traverse the cytosol of several hepatocytes before invading one by forming a parasitophorous vacuole [
20]. The host responds to the traveling parasites by secreting HGF, which renders hepatocytes susceptible to infection [
10]. HGF is known to act as an autocrine cytokine in various cancer cells and precancerous cell types [
21,
22], but it is not normally expressed by hepatocytes or by most other MET-expressing epithelial cells. However, the mechanism of HGF regulation in mammalians is not fully understood.
Moreover, clarifying transcription regulation in
Plasmodium genome is onerous because of the genome's abnormal AT-richness [
23,
24]. Nonetheless,
Plasmodium genes are organized similarly to those in eukaryotes and are also monocistronically transcribed [
25-
27], implying the presence of regulatory sequence elements flanking the coding regions [
28,
29]. For example, the
Pf multidrug resistance gene upstream region can be induced at the level of transcription by antimalarial drugs in chloroquine sensitive parasites [
30]. Our results obtained from upstream transcriptional activities of
Pf-Sub2 demonstrate that the proximal promoter of
Pf-Sub2 is influenced by the HGF. 5'UTR were analyzed for transcriptional elements of which 14 TATA boxes and other elements,
e.g., one Oct-1 and c-Myb were observed. Deletion mutants of 5'UTR of
Pf-Sub2 were constructed and -1,422/+12 UTR exhibited the strongest promoter activity. To understand the interaction of HGF and 5'UTR of
Pf-Sub2, 2 vectors of the molecules are cloned and co-transfected into cells. HGF expression was suppressed by 5'untranslated region of the
Pf-Sub2 promoter. In these results, -1,422 to +12 UTR of
Pf-Sub2 was applied and contained all transcriptional elements in
Table 1. Therefore, further study will be performed what positions of -1,422 to +12 UTR are important to inhibit the HGF expression. Although the effect of HGF on malaria gene expression in cultured cells may be indirect or atypical, the present results indicate the feasibility of regulation of gene(s) important in malaria invasion through host-parasite interaction. In fact, both recombination in subtelomeric regions and chromosome internal rearrangements under host pressure have proven that the parasite can interact with its vertebrate host [
31-
33]. Specific upstream regions are important in the activity of the protease. Future studies will be aimed at elucidating the details of the role of HGF in malaria sensitivity/resistance and how Sub2 gene expression is influenced by the host HGF signaling cascade.
ACKNOWLEDGEMENTS
This research was supported by Wonkwang University in 2009.
References
- 1. Kooij TW, Janse CJ, Waters AP. Plasmodium post-genomics: better the bug you know? Nat Rev Microbiol 2006;4:344-357.
- 2. Fortin A, Stevenson MM, Gros P. Complex genetic control of susceptibility to malaria in mice. Genes Immun 2002;3:177-186.
- 3. Griffith JW, O'Connor C, Bernard K, Town T, Goldstein DR, Bucala R. Toll-like receptor modulation of murine cerebral malaria is dependent on the genetic background of the host. J Infect Dis 2007;196:1553-1564.
- 4. Millington OR, Gibson VB, Rush CM, Zinselmeyer BH, Phillips RS, Garside P, et al. Malaria impairs T cell clustering and immune priming despite normal signal 1 from dendritic cells. PLoS Pathog 2007;3:1380-1387.
- 5. Min-Oo G, Fortin A, Pitari G, Tam M, Stevenson MM, Gros P. Complex genetic control of susceptibility to malaria: positional cloning of the Char 9 locus. J Exp Med 2007;204:511-524.
- 6. Burt RA. Genetics of host response to malaria. Int J Parasitol 1999;29:973-979.
- 7. Mikolajczak SA, Kappe SH. A clash to conquer: the malaria parasite liver infection. Mol Microbiol 2006;62:1499-1506.
- 8. Morosan S, Hez-Deroubaix S, Lunel F, Renia L, Giannini C, Van RN, et al. Liver-Stage Development of Plasmodium falciparum in a Humanized Mouse Model. J Infect Dis 2006;193:996-1004.
- 9. Prudêncio M, Rodriguez A, Mota MM. The silent path to thousands of merozoites: the Plasmodium liver stage. Nat Rev Microbiol 2006;4:849-856.
- 10. Carrolo M, Giordano S, Cabrita-Santos L, Corso S, Vigário AM, Silva S, et al. Hepatocyte growth factor and its receptor are required for malaria infection. Nat Med 2003;9:1363-1369.
- 11. Adachi K, Tsutsui H, Kashiwamura S, Seki E, Nakano H, Takeuchi O, et al. Plasmodium berghei infection in mice induces liver injury by an IL-12- and Toll-like receptor/myeloid differentiation factor 88-dependent mechanism. J Immunol 2001;167:5928-5934.
- 12. Joshi YK, Tandon BN, Acharya SK, Babu S, Tandon M. Acute hepatic failure due to Plasmodium falciparum liver injury. Liver 1986;6:357-360.
- 13. Yoshimoto T, Takahama Y, Wang CR, Yoneto T, Waki S, Nariuchi H. A pathogenic role of IL-12 in blood-stage murine malaria lethal strain Plasmodium berghei NK65 infection. J Immunol 1998;160:5500-5505.
- 14. Chisari FV, Ferrari C. Hepatitis B virus immunopathogenesis. Annu Rev Immunol 1995;13:29-60.
- 15. Goljan J, Nahorski W, Felczak-Korzybska I, Górski J, Myjak P. Liver injury in the course of malaria. Int Marit Health 2000;51:30-39.
- 16. Howell SA, Well I, Fleck SL, Kettleborough C, Collins CR, Blackman MJ. A single malaria merozoite serine protease mediates shedding of multiple surface proteins by juxtamembrane cleavage. J Biol Chem 2003;278:23890-23898.
- 17. Harris PK, Yeoh S, Dluzewski AR, O'Donnell RA, Withers-Martinez C, Hackett F, et al. Molecular identification of a malaria merozoite surface sheddase. PLoS Pathog 2005;1:241-251.
- 18. O'Donnell RA, Hackett F, Howell SA, Treeck M, Struck N, Krnajski Z, et al. Intramembrane proteolysis mediates shedding of a key adhesin during erythrocyte invasion by the malaria parasite. J Cell Biol 2006;174:1023-1033.
- 19. Uzureau P, Barale JC, Janse CJ, Waters AP, Breton CB. Gene targeting demonstrates that the Plasmodium berghei subtilisin PbSUB2 is essential for red cell invasion and reveals spontaneous genetic recombination events. Cell Microbiol 2004;6:65-78.
- 20. Mota MM, Pradel G, Vanderberg JP, Hafalla JCR, Frevert U, Nussenzweig RS, et al. Migration of Plasmodium Sporozoites Through Cells Before Infection. Science 2001;291:141-144.
- 21. Rong S, Segal S, Anver M, Resau JH, Van Woude GF. Invasiveness and metastasis of NIH 3T3 cells induced by Met-hepatocyte growth factor/scatter factor autocrine stimulation. Proc Natl Acad Sci USA 1994;91:4731-4735.
- 22. Jeffers M, Rong S, Ariver M, Vande Woude GF. Autocrine hepatocyte growth factor/scatter factor-Met signaling induces transformation and the invasive/metastatic phenotype in C127 cells. Oncogene 1996;13:853-856.
- 23. van Noort V, Huynen MA. Combinatorial gene regulation in Plasmodium falciparum. Trends Genet 2006;22:73-78.
- 24. Gunasekera AM, Myrick A, Militello KT, Sims JS, Dong CK, Gierahn T, et al. Regulatory motifs uncovered among gene expression clusters in Plasmodium falciparum. Mol Biochem Parasitol 2007;153:19-30.
- 25. Lanzer M, Wertheimer SP, de Bruin D, Ravetch JV. Plasmodium: control of gene expression in malaria parasites. Exp Parasitol 1993;77:121-128.
- 26. Wickham ME, Thompson JK, Cowman AF. Characterisation of the merozoite surface protein-2 promoter using stable and transient transfection in Plasmodium falciparum. Mol Biochem Parasitol 2003;129:147-156.
- 27. Ruvalcaba-Salazar OK, del Carmen Ramírez-Estudillo M, Montiel-Condado D, Recillas-Targa F, Vargas M, Hernández-Rivas R. Recombinant and native Plasmodium falciparum TATA-binding-protein binds to a specific TATA box element in promoter regions. Mol Biochem Parasitol 2005;140:183-196.
- 28. Crabb BS, Cowman AF. Characterization of promoters and stable transfection by homologous and nonhomologous recombination in Plasmodium falciparum. Proc Natl Acad Sci 1996;93:7289-7294.
- 29. Pace T, Olivieri A, Sanchez M, Albanesi V, Picci L, Siden KI, et al. Set regulation in asexual and sexual Plasmodium parasites reveals a novel mechanism of stage-specific expression. Mol Microbiol 2006;60:870-882.
- 30. Myrick A, Munasinghe A, Patankar S, Wirth DF. Mapping of the Plasmodium falciparum multidrug resistance gene upstream region, and evidence of induction of transcript levels by antimalarial drugs in chloroquine sensitive parasites. Mol Microbiol 2003;49:671-683.
- 31. Calderwood MS, Gannoun-Zaki L, Wellems TE, Deitsch KW. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J Biol Chem 2003;278:34125-34132.
- 32. Voss TS, Healer J, Marty AJ, Duffy MF, Thompson JK, Beeson JG, et al. A var gene promoter controls allelic exclusion of virulence genes in Plasmodium falciparum malaria. Nature 2006;439:1004-1008.
- 33. Duffy MF, Tham WH. Transcription and coregulation of multigene families in Plasmodium falciparum. Trends Parasitol 2007;23:183-186.
Fig. 1The promoter activity analysis of pGL3-Basic-Pf-Sub2 5'UTR vector. The promoter activities of 4 different pGL3-Basic-Pf-Sub2 5'UTR constructs were evaluated by luciferase expression (Relative Luciferase Unit, RLU). β-galactosidase activity was used to normalize transfection efficiency.
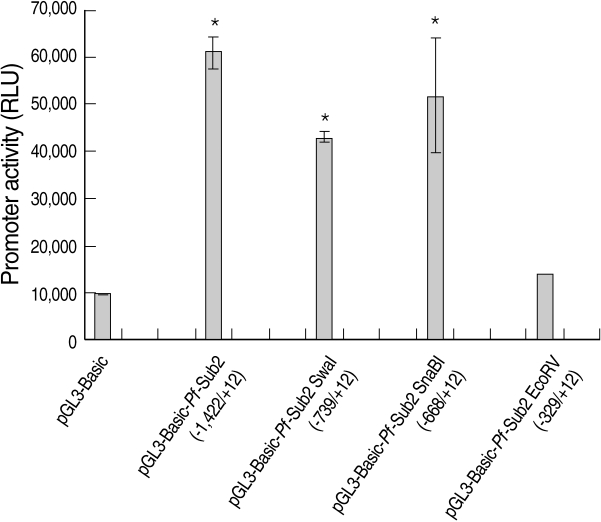
Fig. 2Construction of pcDNA3 HA HGF vector and HGF-expression. HGF was inserted into pcDNA3 HA vector using BamHI and XhoI restriction enzyme sites. DNA fragments were observed for HGF cloning by restriction enzymes digestion (A) Lane 1, HindIII; lane 2, HindIII + XbaI. (B) Shows the expression of 25-kDa-sized HGF. The diagram of pcDNA3 HA HGF vector of about 7.4 kb which contains restriction enzyme sites is provided as supplementary data.

Fig. 3Influence of HGF onto the promoter activity of pGL3-Basic-Pf-Sub2 5'UTR vector. pGL3-Basic-Pf-Sub2 5'UTR (-1,422/+12) and pcDNA3 HA HGF vector were co-transfected into 293T cells. Its promoter activity was estimated by luciferase expression (RLU). β-galactosidase activity was used to normalize transfection efficiency.

Table 1.Putative transcriptional elements in upstream region -1,453 bp of Pf-Sub2
Table 1.
|
Transcriptional elements of Pf- Sub2 upstream 1,453 bp |
Total number |
Trans position |
Cis position |
|
TATA |
14 |
132, 169, 896, 913, 1,074, 1,121, 1,204, 1,228 |
88, 103, 228, 252, 587, 1,204 |
|
XFD-3 |
4 |
140, 446 |
294, 1,315 |
|
USF |
4 |
117, 1,236 |
1,150, 1,234 |
|
c/EBP |
4 |
782, 1,136 |
289, 1,235 |
|
AP-1 |
2 |
1,212 |
723 |
|
Oct-1 |
1 |
- |
532 |
|
c-Myb |
1 |
346 |
- |