Abstract
The onset, severity, and ultimate outcome of malaria infection are influenced by parasite-expressed virulence factors as well as by individual host responses to these determinants. In both humans and mice, liver injury follows parasite entry, persisting to the erythrocytic stage in the case of infection with the fatal strain of Plasmodium falciparum. Hepatic nuclear factor (HNF)-1α is a master regulator of not only the liver damage and adaptive responses but also diverse metabolic functions. In this study, we analyzed the expression of host HNF-1α in relation to malaria infection and evaluated its interaction with the 5'-untranslated region of subtilisin-like protease 2 (subtilase, Sub2). Recombinant human HNF-1α expressed by a lentiviral vector (LV HNF-1α) was introduced into mice. Interestingly, differences in the activity of the 5'-untranslated region of the Pf-Sub2 promoter were detected in 293T cells, and LV HNF-1α was observed to influence promoter activity, suggesting that host HNF-1α interacts with the Sub2 gene.
-
Key words: Plasmodium falciparum, hepatic nuclear factor-1α, subtilisin-like protease 2, lentiviral vector
Malaria, which is caused by infection with the hemoprotozoan parasite
Plasmodium, afflicts approximately 500 million people each year, leading to 2.7 million deaths annually worldwide [
1]. Malaria infections result in diverse clinical outcomes, presumably because of the dynamic relationship between parasite-expressed virulence factors and individual host responses to these determinants [
2]. The last few years have seen a rapid growth in the number of reported genetic associations with host susceptibility and resistance to malaria; these are mainly variants in erythrocytes, cytoadherence factors, immune system, and inflammatory genes [
3-
5]. However, the molecular basis of malaria infection remains incompletely defined. The identification of genetic factors involved in malaria susceptibility and resistance in both the host and the parasite would lead not only to a greater understanding of this complex disease but also potentially to development of effective medical interventions [
2,
6].
Interactions between
Plasmodium and its host hepatocytic factors are of great research interest [
7-
9]. On one hand, it is well established that malaria sporozoites exploit signaling via the hepatocyte growth factor (HGF) and its receptor MET during host invasion [
10]. On the other hand, malaria is a life cycle-specific disease that includes liver injury at the erythrocytic stage of the parasite [
11]. Indeed, both humans and mice suffer from liver injury after infection with the fatal strains of
Plasmodium falciparum (
Pf) and
Plasmodium berghei (
Pb) [
12,
13]. Hepatic nuclear factor (HNF)-1α, a liver-specific transcription factor also designated as LF-B1 or TCF-1, is a critical component of the regulatory network, affecting diverse metabolic functions in the liver, pancreatic islets, intestine, and kidneys. The gene encoding HNF-1α maps to chromosome 12q34 and functions as a dimer [
14]. HNF-1α plays an important role in transactivation of hepatic genes during adaptive responses to liver injury [
15,
16]. Additionally, mutation in the gene is associated with maturity-onset diabetes of the young type III (MODY3) [
16,
17], indicating the important biological role of HNF-1α in glucose metabolism. In addition, HNF-1α interacts with other transactivators to regulate the chromatin structure of the gene encoding the serine protease inhibitor, serpin [
18].
P. falciparum subtilisin-like protease 2 (subtilase, Sub2) is an essential integral membrane serine protease in both
Pf and
Pb. It functions in shedding of the ectodomain components of 2 surface proteins, membrane surface protein 1 (MSP1) and apical membrane antigen 1 (AMA1), upon host invasion [
19,
20]. The importance of the Sub2 gene is evidenced by the fact that Sub2 knock-out parasites cannot be recovered [
21].
In this study, to understand HNF-1α interactions with the malaria antigen Sub2, the activity of the Pf-Sub2 promoter was estimated. We cloned the 5'-untranslated region (UTR) of Pf-Sub2 from Pf genomic DNA (gDNA) and analyzed its transcriptional activity when exogenous HNF-1α was over-expressed in 293T cells.
Pf 3D7 strain was obtained from Malaria Research and Reference Reagent Resource Center (MR4; Manassas, Virginia, USA;
http://www.mr4.org).
Pf 3D7 culture conditions were set according to the MR4 standard protocol. Parasitemia was calculated by counting at least 1,000 cells following staining with Diff Quik (Sysmex, Kobe, Japan). The parasitemia of about 10% to 30% was continuously maintained.
Cell culture media and components, including fetal bovine serum (FBS) were obtained from Invitrogen (Carlsbad, California, USA). All cells were purchased from the American Type Culture Collection (ATCC, Rockville, Maryland, USA) and were routinely cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS. HeLa and 293T cells were used for the experiments.
All plasmids were purified using JETstar2.0 mini and maxi-prep Kit (GenoMed, St. Louis, Missouri, USA) according to the manufacturer's instructions. Pf 3D7 small-scale gDNA isolation followed the MR4 protocol. The HNF-1α cDNA clone was obtained from Open Biosystems (Accession No: BC104908; Rockford, Illinois, USA) and was amplified by PCR with primers F (5'-ATAGAA TTCGCAGCCGAGCCATGGTTTCTA-3'; EcoRI site underlined) and R (5'-ATACTCGAGTGCCGTGGTTACTGGGAGGAAG-3'; XhoI site underlined). The amplified fragment was inserted into pCDNA3-Flag-HA (Addgene, Cambridge, Massachusetts, USA) by EcoRI and XhoI ligation. Lentivirus (LV) HNF-1α was constructed by cutting the LV plasmid backbone with BamHI and XbaI to expel the resident luciferase gene, which was replaced with a HNF-1α cDNA insert cut from pCDNA3-HNF-1α-Flag-HA. A construct for luciferase activity was based on a pGL3-Basic vector (Promega, Madison, Wisconsin, USA). Two different 5'-upstream sequences flanking the Pf-Sub2 gene were amplified from Pf 3D7 gDNA with primers. The primers used for -2,567/+32 UTR were F (5'-ATAGGTACCTAGAATAGAATAAAATAAATAACC-3'; KpnI site underlined) and R (5'-ATAAAGCTTATCAAGGAAACCACATAA-3'; HindIII site underlined). The primers used for -1,422/+12 UTR were F (5'-AAGGTACCATGGTATGAGTTCTTTATA-3'; KpnI site underlined) and R (5'-ATGCTAGCAATATTCAGCATTATACGGA-3'; NheI site underlined). PCR was performed using a TGRADIENT thermocycler (Biometra, Goettingen, Germany). The amplified UTR sequences were cloned into a pMDT vector (Takara Bio, Shiga, Japan) and subcloned into a pGL3-Basic vector with KpnI and NheI/HindIII sticky end ligation, generating the pGL3-Basic-Pf-Sub2 5'-UTR construct. The other upstream inserts were cloned into a pGL3-Basic vector by blunt restriction sites (EcoRV, SnaBI and SwaI) located in the 1.5-kb region upstream of the luciferase gene. The internal control was pSV-β-galactosidase (Promega). All clones were analyzed by restriction mapping and were sequenced by a 3730XL DNA sequencer (Macrogen, Seoul, Korea).
A human immunodeficiency virus (HIV)-1-based LV plasmid system, including the LV backbone with luciferase gene flanked at the 5'-end by cytomegalovirus vector (CMV) promoter, pCMV dR8.91 packaging construct, and pMDG2 envelope plasmid, were kindly provided by Dr. Didier Trono. The pseudo-typed LV was generated by co-transfecting these plasmids into 293T cells using the calcium phosphate method [
22]. LV was concentrated by overlaying the viral supernatant on a 20% sucrose cushion and centrifuging at 12,000 rpm for 2 hr at 4℃ in a Beckman SW28 swing bucket rotor. Aliquots of the concentrated virus were stored at -80℃. LV preparation quality was determined by using serial dilutions of the concentrated frozen viral stock to infect HeLa cells contained in 6-well plates (NUNC, Roskilde, Denmark). After 48 hr incubation, luciferase activity of cells infected with 10 µL of a concentrated virus suspension was determined. The recombinant HNF-1α was analyzed with an anti-HNF-1α antibody by western blotting as described below.
Five-week-old mice were injected with LV HNF-1α through the tail vein. The 200 µl injected volume varied in potency from a low dose consisting of 20 µl concentrated LV HNF-1α and 180 µl of phosphate-buffered saline (PBS) to a high dose consisting of 200 µl of concentrated LV HNF-1α. Blank control mice were injected with the same volume of PBS, and mimic control mice were injected with the LV backbone lacking HNF-1α. DNA isolated from liver tissue from LV HNF-1α-infected mice was subjected to reverse transcription (RT)-PCR with human HNF-1α-specific primers that contained the maximum number of critical mismatches with the homologous mouse sequence (F: 5'-CCCTGGCCTGCCTCCACCTG-3' and R: 5'-GGGTCACATGGCTCTGCACAGGT-3').
Cells were harvested and lysed with RIPA buffer (Pierce, Rockford, Illinois, USA) with freshly added protease inhibitor cocktail (Sigma-Aldrich, St. Louis, Missouri, USA). One hundred micrograms of cell lysate was diluted with a loading buffer that contained a reducing agent, and proteins were separated on 12% SDS-PAGE and transferred to a polyvinylidene fluoride membrane. Immunoblotting was performed with a 1:200 dilution of rabbit polyclonal anti-HNF-1α antibody (SC10791; Santa Cruz Biotechnology, Santa Cruz, California, USA) in 3% bovine serum albumin (BSA). The secondary antibody, polyclonal goat anti-rabbit IgG conjugated with horseradish peroxidase (HRP) (Dako, Glostrup, Denmark), was used at a 1:1,000 dilution in PBS. For proliferating cell nuclear antigen (PCNA) quantification, anti-PCNA/HRP monoclonal antibody was used (Dako).
Cells were transfected with 2.5 µg of the reporter constructs and 0.5 µg of pCMV-β-galactosidase by a standard calcium phosphate co-precipitation method in 6-well plates. For co-transfection, 1.5 µg of the reporter constructs and 0.5 µg of pCMV-β-galactosidase were co-transfected with different amounts of pCDNA-HNF-1α-Flag-HA (0.5-4.5 µg). Each sample was analyzed in duplicate and each experiment was repeated twice. Luciferase and β-galactosidase activities were measured using a model LB96V microplate luminometer (Berthold Technologies, Bad Wildbad, Germany) according to the manufacturer's instructions. Luciferase activity was normalized with respect to β-galactosidase activity to correct deviation due to transfection efficiencies and cell numbers.
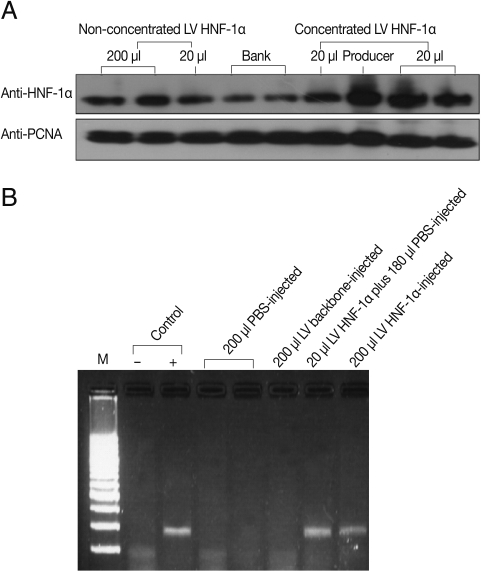
HNF-1α gene expression was observed both in vitro and in vivo. To determine LV HNF-1α production, 20 µl and 200 µl of non-concentrated (approximately 1×10
7 transducing units (TU)/mL) or 20 µl of concentrated (approximately 10
9 TU/mL) LV HNF-1α was used to infect cultured HeLa cells. After 48 hr, western blot analyses using monoclonal anti-HNF-1α antibody revealed strong gene expression in cells infected with 20 µl of concentrated LV HNF-1α (
Fig. 1A). Six days following injection of LV backbone and LV HNF-1α into mice, luciferase gene expression was detected in peripheral blood (data not shown). To confirm the expression of human HNF-1α gene in mice, RT-PCR was performed with mice liver tissues. Human HNF-1α was amplified in mice infected with LV HNF-1α but not in mice injected with the mimic LV backbone or PBS (
Fig. 1B).
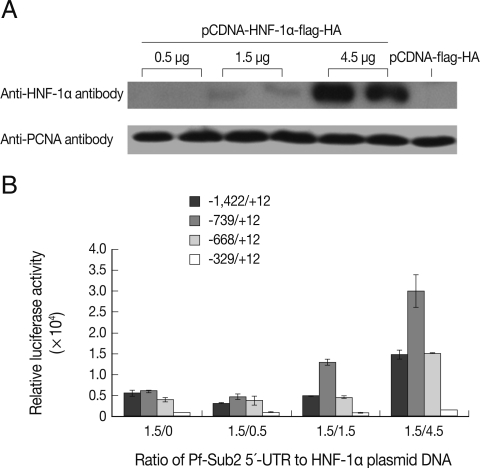
To analyze the influence of HNF-1α on the transcriptional activity of 5'-UTR of
Pf-Sub2 in vitro, co-transfection of pGL3-Basic-
Pf-Sub2 5'-UTR with different amounts of HNF-1α was performed in 293T cells. The exogenous HNF-1α expression level of pCDNA-HNF-1α-Flag-HA in transfected 293T cells was ascertained by western blotting with anti-HNF-1α antibody (
Fig. 2A). High HNF-1α expression was observed using 4.5 µg of the pCDNA-HNF-1α-Flag-HA transfection vector. Except for the -329/+12 UTR, the 5'-UTRs showed high and distinct transcriptional luciferase activities (
Fig. 2B). As the amount of pCDNA-HNF-1α-Flag-HA vector increased from 0.5 to 4.5 µg, luciferase activity was increased. Although the -1,422/+12 UTR did not drive the luciferase activity in a similar manner as apparent in
Fig. 2A, a significant difference was seen with 1.5 µg of pGL3-Basic-
Pf-Sub2 5'-UTR/4.5 µg of pCDNA-HNF-1α-flag-HA as compared with only 1.5 µg of pGL3-Basic-
Pf-Sub2 5'-UTR.
HNF-1α is a critical regulator of diverse metabolic functions in the liver, pancreatic islets, intestine, and kidneys. It is essential for glucose, bile acid, and plasma cholesterol metabolism [
15,
16,
23]. However, further details pertaining to HNF-1α function remain to be resolved. HNF-1α not only physically interacts with basic transcriptional machinery, such as histone acetyltransferases (HATs), CREB-binding protein (CBP), and p300/CBP-associated factor (P/CAF) [
24], but also plays an important role in modulation of chromatin structure, as in the case of hyperacetylation of the serpin gene [
18,
25]. Hence, exogenous HNF-1α signaling cascades might influence basic regulatory networks, metabolic states, and immune system functioning of the host, thereby indirectly influencing the infecting
Plasmodium. Since the molecular and cellular factors mediating differences in malaria virulence and severity are poorly understood [
26], the present study might lay the groundwork for research investigating different natural outcomes of malaria infections.
The mechanism of HNF-1α regulation in mammals is not fully understood. Moreover, clarifying transcription regulation in the
Plasmodium genome is onerous because of the genome's abnormal AT-richness [
27,
28]. Nonetheless,
Plasmodium genes are organized similarly to those in eukaryotes and are also monocistronically transcribed [
29-
31], implying the presence of regulatory sequence elements flanking the coding regions [
32,
33]. For example, the region upstream to the
Pf multidrug resistance gene can be induced at the transcriptional level by antimalarial drugs in chloroquine-sensitive parasites [
34]. Our results obtained from upstream transcriptional activities of
Pf-Sub2 demonstrate that the proximal promoter of
Pf-Sub2 is influenced by exogenous HNF-1α. Although the effect of HNF-1α on malaria gene expression in cultured cells may be indirect or atypical, the present results indicate the feasibility of regulation of genes important for malaria invasion via host-parasite interaction.
The present study has shown that exogenous human HNF-1α expression is involved in host response to malaria infection. Interestingly, host HNF-1α interacts at the mRNA level with the parasite serine protease Sub2, which is important in host cell invasion by the malarial parasite. Specific upstream regions are important in the activity of the protease. Taken together, these data suggest that human HNF-1α interacts with the promoter regions of the Sub2 gene. Future studies will be aimed at elucidating the role of HNF-1α in malaria sensitivity/resistance and how Sub2 gene expression is influenced by the host HNF-1α signaling cascade.
ACKNOWLEDGMENTS
We thank Dr. Zhang Sizhong for his constructive suggestions, Dr. Didier Trono for kindly providing the lentiviral vector system, and Dr. Peter Mackenzie for the HNF-1α plasmid construct. This research was supported by Wonkwang University in 2010.
References
- 1. Kooij TW, Janse CJ, Waters AP. Plasmodium post-genomics: better the bug you know? Nat Rev Microbiol 2006;4:344-357.
- 2. Fortin A, Stevenson MM, Gros P. Complex genetic control of susceptibility to malaria in mice. Genes Immun 2002;3:177-186.
- 3. Griffith JW, O'Connor C, Bernard K, Town T, Goldstein DR, Bucala R. Toll-like receptor modulation of murine cerebral malaria is dependent on the genetic background of the host. J Infect Dis 2007;196:1553-1564.
- 4. Millington OR, Gibson VB, Rush CM, Zinselmeyer BH, Phillips RS, Garside P, Brewer JM. Malaria impairs T cell clustering and immune priming despite normal signal 1 from dendritic cells. PLoS Pathog 2007;3:1380-1387.
- 5. Min-Oo G, Fortin A, Pitari G, Tam M, Stevenson MM, Gros P. Complex genetic control of susceptibility to malaria: positional cloning of the Char9 locus. J Exp Med 2007;204:511-524.
- 6. Burt RA. Genetics of host response to malaria. Int J Parasitol 1999;29:973-979.
- 7. Mikolajczak SA, Kappe SH. A clash to conquer: The malaria parasite liver infection. Mol Microbiol 2006;62:1499-1506.
- 8. Morosan S, Hez-Deroubaix S, Lunel F, Renia L, Giannini C, Van Rooijen N, Battaglia S, Blanc C, Eling W, Sauerwein R, Hannoun L, Belghiti J, Brechot C, Kremsdorf D, Druilhe P. Liver-stage development of Plasmodium falciparum, in a humanized mouse model. J Infect Dis 2006;193:996-1004.
- 9. Prudêncio M, Rodriguez A, Mota MM. The silent path to thousands of merozoites: The Plasmodium liver stage. Nat Rev Microbiol 2006;4:849-856.
- 10. Carrolo M, Giordano S, Cabrita-Santos L, Corso S, Vigário AM, Silva S, Leirião P, Carapau D, Armas-Portela R, Comoglio PM, Rodriguez A, Mota MM. Hepatocyte growth factor and its receptor are required for malaria infection. Nat Med 2003;9:1363-1369.
- 11. Adachi K, Tsutsui H, Kashiwamura S, Seki E, Nakano H, Takeuchi O, Takeda K, Okumura K, Van Kaer L, Okamura H, Akira S, Nakanishi K. Plasmodium berghei infection in mice induces liver injury by an IL-12- and toll-like receptor/myeloid differentiation factor 88-dependent mechanism. J Immunol 2001;167:5928-5934.
- 12. Joshi YK, Tandon BN, Acharya SK, Babu S, Tandon M. Acute hepatic failure due to Plasmodium falciparum liver injury. Liver 1986;6:357-360.
- 13. Yoshimoto T, Takahama Y, Wang CR, Yoneto T, Waki S, Nariuchi H. A pathogenic role of IL-12 in blood-stage murine malaria lethal strain Plasmodium berghei NK65 infection. J Immunol 1998;160:5500-5505.
- 14. Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, Talianidis I. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev 2006;20:2293-2305.
- 15. Cerf ME. Transcription factors regulating beta-cell function. Eur J Endocrinol 2006;155:671-679.
- 16. Gautier-Stein A, Zitoun C, Lalli E, Mithieux G, Rajas F. Transcriptional regulation of the glucose-6-phosphatase gene by cAMP/vasoactive intestinal peptide in the intestine: Role of HNF4alpha, CREM, HNF1alpha, and C/EBPalpha. J Biol Chem 2006;281:31268-31278.
- 17. Jain S, Li Y, Patil S, Kumar A. HNF-1alpha plays an important role in IL-6-induced expression of the human angiotensinogen gene. Am J Physiol Cell Physiol 2007;293:C401-C410.
- 18. Rollini P, Fournier RE. The HNF-4/HNF-1alpha transactivation cascade regulates gene activity and chromatin structure of the human serine protease inhibitor gene cluster at 14q32.1. Proc Natl Acad Sci U S A 1999;96:10308-10313.
- 19. Howell SA, Well I, Fleck SL, Kettleborough C, Collins CR, Blackman MJ. A single malaria merozoite serine protease mediates shedding of multiple surface proteins by juxtamembrane cleavage. J Biol Chem 2003;278:23890-23898.
- 20. Harris PK, Yeoh S, Dluzewski AR, O'Donnell RA, Withers-Martinez C, Hackett F, Bannister LH, Mitchell GH, Blackman MJ. Molecular identification of a malaria merozoite surface sheddase. PLoS Pathog 2005;1:241-251.
- 21. Uzureau P, Barale JC, Janse CJ, Waters AP, Breton CB. Gene targeting demonstrates that the Plasmodium berghei subtilisin PbSUB2 is essential for red cell invasion and reveals spontaneous genetic recombination events. Cell Microbiol 2004;6:65-78.
- 22. Wigler M, Pellicer A, Silverstein S, Axel R. Biochemical transfer of single-copy eucaryotic genes using total cellular DNA as donor. Cell 1978;14:725-731.
- 23. Shih DQ, Bussen M, Sehayek E, Ananthanarayanan M, Shneider BL, Suchy FJ, Shefer S, Bollileni JS, Gonzalez FJ, Breslow JL, Stoffel M. Hepatocyte nuclear factor-1α is an essential regulator of bile acid and plasma cholesterol metabolism. Nat Genet 2001;27:375-382.
- 24. Soutoglou E, Papafotiou G, Katrakili N, Talianidis I. Transcriptional activation by hepatocyte nuclear factor-1 requires synergism between multiple coactivator proteins. J Biol Chem 2000;275:12515-12520.
- 25. Baxter EW, Cummings WJ, Fournier RE. Formation of a large, complex domain of histone hyperacetylation at human 14q32.1 requires the serpin locus control region. Nucleic Acids Res 2005;33:3313-3322.
- 26. Wykes MN, Liu XQ, Beattie L, Stanisic DI, Stacey KJ, Smyth MJ, Thomas R, Good MF. Plasmodium strain determines dendritic cell function essential for survival from malaria. PLoS Pathog 2007;3:e96.
- 27. van Noort V, Huynen MA. Combinatorial gene regulation in Plasmodium falciparum. Trends Genet 2006;22:73-78.
- 28. Gunasekera AM, Myrick A, Militello KT, Sims JS, Dong CK, Gierahn T, Le Roch K, Winzeler E, Wirth DF. Regulatory motifs uncovered among gene expression clusters in Plasmodium falciparum. Mol Biochem Parasitol 2007;153:19-30.
- 29. Lanzer M, Wertheimer SP, de Bruin D, Ravetch JV. Plasmodium: control of gene expression in malaria parasites. Exp Parasitol 1993;77:121-128.
- 30. Wickham ME, Thompson JK, Cowman AF. Characterisation of the merozoite surface protein-2 promoter using stable and transient transfection in Plasmodium falciparum. Mol Biochem Parasitol 2003;129:147-156.
- 31. Ruvalcaba-Salazar OK, del Carmen Ramírez-Estudillo M, Montiel-Condado D, Recillas-Targa F, Vargas M, Hernández-Rivas R. Recombinant and native Plasmodium falciparum TATA-binding-protein binds to a specific TATA box element in promoter regions. Mol Biochem Parasitol 2005;140:183-196.
- 32. Crabb BS, Cowman AF. Characterization of promoters and stable transfection by homologous and nonhomologous recombination in Plasmodium falciparum. Proc Natl Acad Sci U S A 1996;93:7289-7294.
- 33. Pace T, Olivieri A, Sanchez M, Albanesi V, Picci L, Siden Kiamos I, Janse CJ, Waters AP, Pizzi E, Ponzi M. Set regulation in asexual and sexual Plasmodium parasites reveals a novel mechanism of stage-specific expression. Mol Microbiol 2006;60:870-882.
- 34. Myrick A, Munasinghe A, Patankar S, Wirth DF. Mapping of the Plasmodium falciparum multidrug resistance gene 5'-upstream region, and evidence of induction of transcript levels by antimalarial drugs in chloroquine sensitive parasites. Mol Microbiol 2003;49:671-683.
Fig. 1Expression of LV HNF-1α. (A) Western blot using an anti-HNF-1α antibody in HeLa cells transduced with concentrated or non-concentrated LV HNF-1α and using an anti-PCNA antibody as a loading control. (B) HNF-1α mRNA expression in transduced mice liver tissues as assessed by RT-PCR.

Fig. 2Co-transfection of Pf-Sub2 UTR constructs with different amounts of exogenous LV HNF-1α in 293T cells. (A) Western blot with an anti-HNF-1α antibody and anti-PCNA as a loading control. (B) Relative luciferase activity of Pf-Sub2 UTR constructs with different amounts of LV HNF-1α (mean±SD).