Abstract
From 2006 to 2010, hospitals in Hanoi treated 10 human patients for dirofilariasis. The worms were collected from parasitic places, and identification of the species was completed by morphology and molecular methods. Ten parasites were recovered either from the conjunctiva (n=9) or subcutaneous tissue (n=1). The parasites were 4.0-12.5 cm in length and 0.5-0.6 mm in width. Morphological observations suggested all parasites as Dirofilaria repens. Three of the 10 parasites (1 from subcutaneous tissue and 2 from eyes) were used for molecular confirmation of the species identification. A portion of the mitochondrial cox1 (461 bp) was amplified and sequenced. Nucleotide and amino acid homologies were 95% and 99-100%, respectively, when compared with D. repens (Italian origin, GenBank AJ271614; DQ358814). This is the first report of eye dirofilariasis and the second report of subcutaneous tissue dirofilariasis due to D. repens in Vietnam.
-
Key words: Dirofilaria repens, case report, eye, subcutaneous tissue, PCR, mitochondrial cox1, Vietnam
INTRODUCTION
Human filariasis is caused by members of the Filaridae family, including species of
Dirofilaria,
Brugia,
Wuchereria,
Onchocerca,
Dipetalonema,
Loa, and
Meningonema; it is transmitted to humans by various kinds of insect vectors [
1]. Dirofilariasis is typically a disease of animals, which can also be easily transmitted to humans by mosquitoes of the genera
Anopheles,
Culex, and
Aedes [
2]. All of these mosquitoes are found in Vietnam. Of the 30 different species of
Dirofilaria,
D. repens and
D. immitis are the 2 most common species that frequently infect humans [
3]. Other
Dirofilaria species have also been reported to infect Vietnamese carnivore species [
4].
There have been over 1,000 cases of dirofilariasis, reported throughout the world, including 300 cases involving the lungs or viscera and over 800 cases involving the subcutaneous tissues or eyes [
1]. Most of these were caused by
D. immitis or
D. repens.
D. immitis is a parasite of dogs and cats and it can occasionally become a causative agent of lung and subcutaneous dirofilariasis in humans.
D. repens can also infect humans and is associated with diseases of various organs, including the conjunctiva, lungs, soft tissues (including the breast), brain, liver, intestine, lymphatic glands, and muscles [
5,
6].
In some cases, identification of
Dirofilaria spp. based only on the morphology is difficult. Therefor, the use of molecular methods, such as PCR, is necessary for effective species identification [
7]. Nuclear and mitochondrial genes are useful for the identification of helminth species, and especially the latter genes have been frequently used for identification of
Dirofilaria spp. [
8-
10].
Given that there has been an increasing number of patients suffering from D. repens infection, further research is required on this newly emerging zoonotic disease as a public health threat in Vietnam. Accurate diagnosis, proper identification, and control measures are therefore needed to control human dirofilariasis in Vietnam.
CASE RECORD
During 2006 to 2010, a total of 9 patients with a swelling mass under their conjunctiva admitted to the National Eye Hospital (NEH), and a patient with a swelling in the subcutaneous tissue admitted to the Military Hospital 108. By surgery, live parasites were collected from these patients and species identification was tried. The total 10 patients, 27-77 years old, were from 4 provinces in the North Vietnam, including Hanoi City (4 patients), Ninh Binh province (3 patients), Ha Nam province (2 patients), and Hung Yen province (1 patient) (
Table 1). Nine of them had similar symptoms, such as a painful, itchy, swollen, and tangible nodule in the eye; 6 cases involved the right eye and 3 involved the left. Another patient, 36-year-old, had a tumor (3×4 cm) in the left subcutaneous tissue, which appeared as a red nodule and was itchy. Surgical biopsies were performed on all patients and living parasites were recovered from each patient.
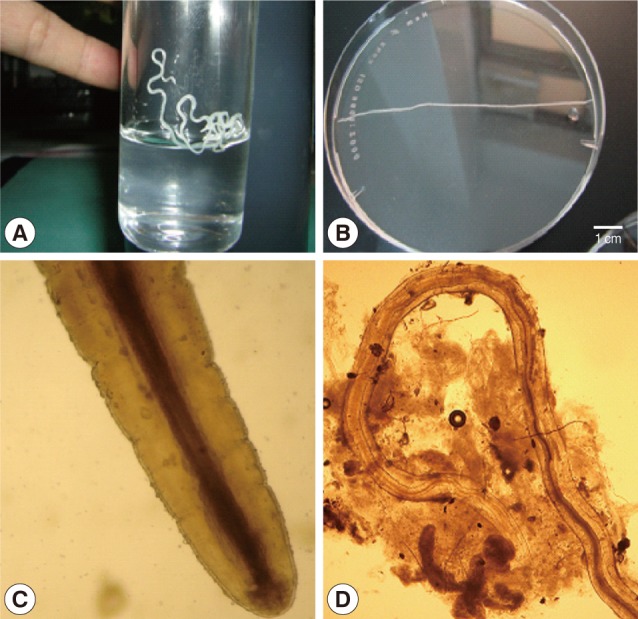
Parasites measured 4.0-12.5 cm in length and 0.5-0.6 mm in width. The worms were identified by the morphology, and pictures were taken (
Fig. 1). Among the worms, 3 were chosen (2 from the conjunctiva and 1 from the subcutaneous tissue), marked as GCA-VN1, GCH-VN2, and GCD-VN3, respectively, and analyzed by molecular methods.
Parasites recovered from the conjunctiva of the eye and subcutaneous tissue were identified as
D. repens on the basis of the morphological keys by Miyazaki in 1991 [
11]. Molecular characterization was conducted as follows: genomic DNA was extracted from individual parasites using a Qiagen genomic DNA extraction Kit (Qiagen, Valencia, California, USA). Extracted genomic DNA was diluted to a working concentration of 100 ng/µl, and 1 µl of this was used in 50 µl PCR reaction volume. PCR amplified a fragment of the cytochrome
c oxidase subunit 1 (
cox1), using the UCO1F1-UCO1R2 primer pairs and additionally as previously described as follows [
9,
12]: UCO1F1: 5'GGTGTTGGTTGAACTTTTTATCCTCC3' and UCO1R2: 5'CCAACCATAAACATATGATGAGCCCA3'.
PCR products purified using a QIAquick Purification Kit (Qiagen) were subjected to direct sequencing using the Big-Dye Terminator Cycle Sequencing technology on an automated sequencer, ABI 3100 Avant Genetic Analyzer (Applied Biosystems, Foster City, California, USA). Sequences were then edited using SeqEdv1.03, aligned using Assembly LIGNv1.9c, and analyzed using the MacVector 8.2 package (Accelrys Inc., San Diego, California, USA). Sequences were searched against the GenBank database, using the NCBI BLAST program (
http://www.ncbi.nlm.nih.gov/blast/Blast.cgi), and approximately 500 bp of the
cox1 of
D. repens from Italy and others sequences were used for comparative purposes. The
cox1 sequences of the Vietnamese
Dirofilaria, including parasites from the conjunctiva (GCA-VN1 and GCH-VN2) and subcutaneous tissue (GCD-VN3), were compared with
D. repens from Italy (ITA1 and ITA2),
D. immitis from Australia (Dimm),
Brugia malayi (Bmal), and
Onchocerca volvulus (Ovol), using GENEDOC2.5 and MEGA3.1 (
Table 2).
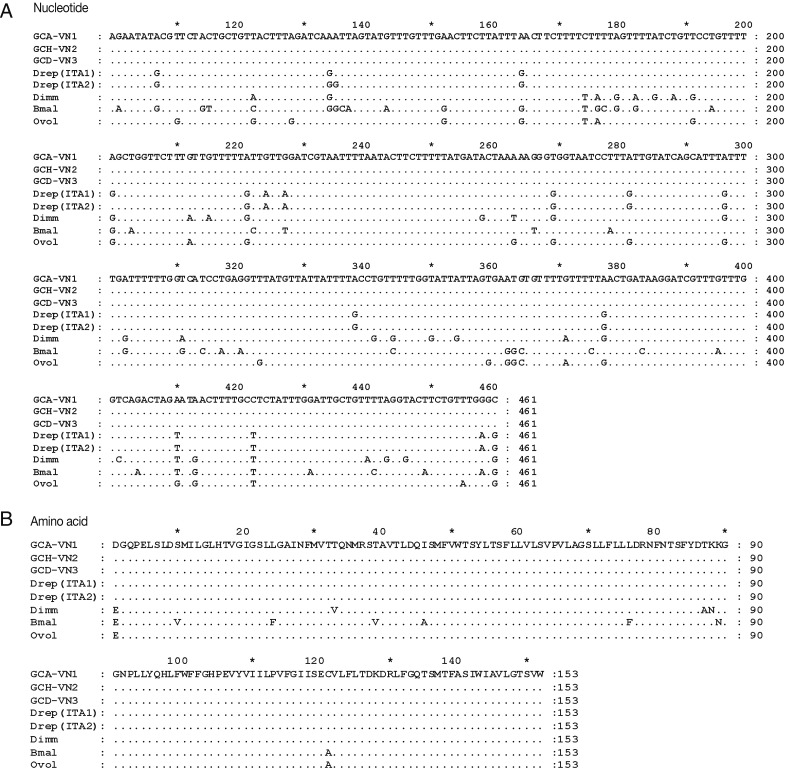
PCR products (500 bp of
cox1) were successfully sequenced, using UCO1F1 and UCO1R2 primers. A portion of
cox1 from the Vietnamese
Dirofilaria
cox1, including 461 nucleotides (A) and 153 amino acids (B), were compared with the
cox1 from Italian
D. repens (Drep [ITA1] and Drep [ITA2], Australian
D. immitis [Dimm],
B. malayi [Bmal], and
O. volvulus [Ovol]) (
Table 3;
Fig. 2).
The 3 portions of the
cox1 sequences of the Vietnamese
Dirofilaria exhibited a 95% nucleotide and 99-100% amino acid identity with the Italian
D. repens (GenBank no. AJ271614 and DQ358814). In contrast, these sequences exhibited a 89% nucleotide and 96% amino acid identity with the Australian
D. immitis; 87% (nucleotide) and 94% (amino acid) similarity with
B. malayi (AF538716); and 91% (nucleotide) and 98% (amino acid) similarity with
O. volvulus (AF015193) (
Table 3;
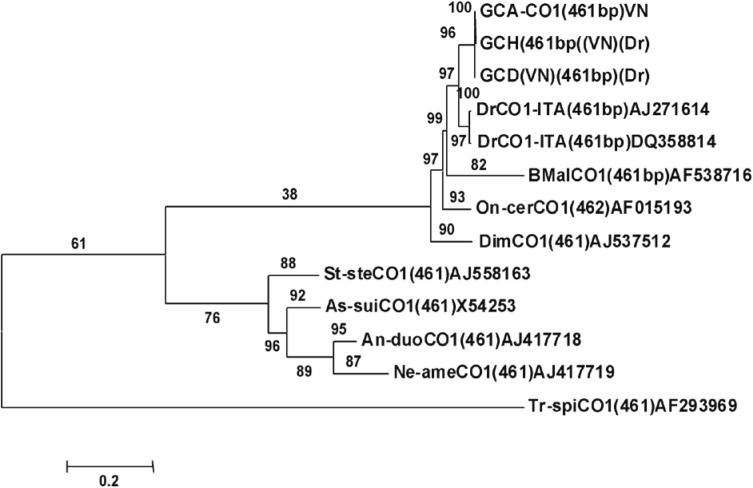
Fig. 2). The phylogenetic analysis results are shown in
Fig. 3. Consequently, we characterized the species of filaria samples from the eye and subcutaneous tissue of patients in Vietnam as
D. repens (Nematoda: Filarioidea).
DISCUSSION
In this study, 10
Dirofilaria worm specimens from humans, including 9 from the conjunctiva and 1 from the subcutaneous tissue, were identified by the morphology and molecular methods as
D. repens. In Vietnam, the filarial worm of this species was first reported from a human conjunctiva in 2008 [
12], and another was reported from the human subcutaneous tissue in 2010 [
13]. This is the 3rd report of human
D. repens infection in Vietnam which involved the conjunctiva or subcutaneous tissue.
This species is parasitic in dogs, cats, and wild animals [
1], and together with
D. immitis it can cause complicated epidemiology in zoonotic diseases. Dirofilariasis is transmitted to humans by mosquitoes, including
Anopheles,
Culex, and
Aedes [
2], and these mosquitoes are common in Vietnam. Feeding dogs and cats are very common in the whole country. Thus, a high risk for human dirofiliasis is existing everywhere in Vietnam.
ACKNOWLEDGMENTS
The authors acknowledge the funds supported from the National Foundation for Science and Technology Development (NAFOSTED) in Vietnam (No. 106.12-2011.13 to Nguyen Van De) and cooperation of researchers from the Hanoi Medical University (HMU), Institute of Biotechnolnotogy (IBT), and the National Eye Hospital (NEH) of Vietnam.
References
- 1. Pampiglione S, Rivasi F. Human dirofilariasis due to Dirofilaria (Nochtiella) repens: An update of world literature from 1995 to 2000. Parassitologia 2000;42:231-254.
- 2. Cancrini G, Scaramozzino P, Gabrielli S, Di Paolo M, Toma L, Romi R. Aedes albopictus and Culex pipiens implicated as natural vectors of Dirofilaria repens in central Italy. J Med Entomol 2007;44:1064-1066.
- 3. Canestri Trotti G, Pampiglione S, Rivasi F. The species of the genus Dirofilaria Railliet & Henry, 1911. Parassitologia 1997;39:369-374.
- 4. Hoa LV, Ty LT. Comparative study of Dirofilaria macacae, Sandground 1933, a parasite of primates, and Dirofilaria repens, Raillet and Henry 1911, a parasite of Vietnamese carnivora. Bull Soc Pathol Exot Filiales 1971;64:347-360.
- 5. Dujic MP, Mitrovic BS, Zec IM. Orbital swelling as a sign of live Dirofilaria repens in subconjunctival tissue. Scand J Infect Dis 2003;35:430-431.
- 6. Raniel Y, Machamudov Z, Garzozi HJ. Subconjunctival infection with Dirofilaria repens. Isr Med Assoc J 2006;8:139.
- 7. Rishniw M, Barr SC, Simpson KW, Frongillo MF, Franz M, Dominguez-Alpizar JL. Discrimination between six species of canine microfilariae by a single polymerase chain reaction. Vet Parasitol 2006;135:303-314.
- 8. Casiraghi M, Anderson TJ, Bandi C, Bazzocchi C, Genchi C. A phylogenetic analysis of filarial nematodes: Comparison with the phylogeny of Wolbachia endosymbionts. Parasitology 2001;122:93-103.
- 9. Hu M, Gasser RB. Mitochondrial genomes of parasitic nematodes-progress and perspectives. Trends Parasitol 2006;22:78-84.
- 10. Le TH, Blair D, McManus DP. Mitochondrial genomes of parasitic flatworms. Trends Parasitol 2002;18:206-213.
- 11. Miyazaki I. An Illustrated Book of Helminthic Zoonoses. 1991, Tokyo, Japan. International Medical Foundation of Japan; pp 422-436 Southeast Asian Medical Information Center.
- 12. De NV, Le TH, Chau HTM, Huan LQ. Human dirofilariasis in the world and the first identification for species in Vietnam. J Pharmaceut Med 2008;5:11-15. (in Vietnamese with English abstract).
- 13. Dang TCT, Nguyen TH, Dung DT, Uga S, Morishima Y, Sugiyama H, Yamasaki H. A human case of subcutaneous dirofilariasis caused by Dirofilaria repens in Vietnam: Histologic and molecular confirmation. Parasitol Res 2010;107:1003-1007.
- 14. Hu M, Gasser RB, Abs El-Osta YG, Chilton NB. Structure and organization of the mitochondrial genome of the canine heartworm, Dirofilaria immitis. Parasitology 2003;127:37-51.
- 15. Ghedin E, Wang S, Spiro D, Caler E, Zhao Q, Crabtree J, Allen JE, Delcher AL, Guiliano DB, Miranda-Saavedra D, Angiuoli SV, Creasy T, Amedeo P, Haas B, El-Sayed NM, Wortman JR, Feldblyum T, Tallon L, Schatz M, Shumway M, Koo H, Salzberg SL, Schobel S, Pertea M, Pop M, White O, Barton GJ, Carlow CK, Crawford MJ, Daub J, Dimmic MW, Estes CF, Foster JM, Ganatra M, Gregory WF, Johnson NM, Jin J, Komuniecki R, Korf I, Kumar S, Laney S, Li BW, Li W, Lindblom TH, Lustigman S, Ma D, Maina CV, Martin DM, McCarter JP, McReynolds L, Mitreva M, Nutman TB, Parkinson J, Peregrín-Alvarez JM, Poole C, Ren Q, Saunders L, Sluder AE, Smith K, Stanke M, Unnasch TR, Ware J, Wei AD, Weil G, Williams DJ, Zhang Y, Williams SA, Fraser-Liggett C, Slatko B, Blaxter ML, Scott AL. Draft genome of the filarial nematode parasite Brugia malayi. Science 2007;317:1756-1760.
- 16. Keddie EM, Higazi T, Unnasch TR. The mitochondrial genome of Onchocerca volvulus: sequence, structure and phylogenetic analysis. Mol Biochem Parasitol 1998;95:111-127.
- 17. Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 2004;101:11030-11035.
Fig. 1
Dirofilaria repens collected from the conjunctiva (A, B) and subcutaneous tissue (C, D) of humans in Vietnam. (A) A female worm from the conjunctiva of a patient. (B) Another specimen from another patient. (C) Anterior end of a worm showing the mouth and esophagus (×100). (D) Posteror part of a female worm extracted from the subcutaneous tissue of a patient (×40).

Fig. 2Comparison of 461 nucleotide (A) and 153 amino acids (B) of portion cox1 mitochondrial genome between Vietnamese Dirofilaria and other species of the family Filaridae, including the Italian Dirofilaria repens (Drep [ITA1] and Drep [ITA2]), Australian D. immitis (Dimm), Brugia malayi (Bmal), and Onchocerca volvulus (Ovol). Note differences between the Vietnamese Dirofilaria (GCA-VN1; GCH-VN2; GCD-VN3) and other species showed by sign nucleotide (or amino acid) of them; mark (.) is similar each other in nucleotide (or amino acid).

Fig. 3The phylogenetic tree based on a portion of
cox1 sequence of
Dirofilaria isolates and nematode strains, including 3 collected in Vietnam. Topology was constructed by MEGA 4.1 using the neighbor-joining method [
17].
Dirofilaria repens from this study and 2 from Italy are shown by the vertical bar. The length of the
cox1 sequence is indicated in brackets. Bootstrap values (%) are indicated in numerals from 1,000 replicates. GCA-VN1, GCH-VN2, and GCD-VN3=Vietnamese
Dirofilaria; Drep (ITA1) and Drep (ITA2)=Italian
Dirofilaria repens (GenBank no. AJ271614 and DQ358814); Dimm:
Dirofilaria immitis (GenBank no. AJ537512); Bmal:
Brugia malayi (GenBank no. AF538716); Ovol:
Onchocerca volvulus (GenBank no. AF015193); St-ste=
Strongyloides stercoralis (GenBank no. AJ558163); As-sui=
Ascaris sum (GenBank no. X54253); An-duo=
Ancylostoma duodenale (GenBank no. AJ417718); Ne-ame=
Neocator americanus (GenBank no.AJ417719); Tr-spi=
Trichinella spiralis (GenBank no. AF293969).

Table 1.Information of worms collected from patients
Table 1.
|
Serial no. cases |
Sex |
Age (years) |
Province |
Parasitic place |
Worm length (cm) |
|
1 |
Female |
50 |
Hanoi |
Right conjunctiva |
4.0 |
|
2 |
Male |
47 |
Hanoi |
Left conjunctiva |
8.0 |
|
3 |
Female |
27 |
Hanoi |
Left conjunctiva |
10.0 |
|
4 |
Male |
49 |
Hanoi |
Right conjunctiva |
5.0 |
|
5 |
Male |
77 |
Ninh Binh |
Right conjunctiva |
11.0 |
|
6 |
Female |
60 |
Ninh Binh |
Right conjunctiva |
15.0 |
|
7 |
Female |
55 |
Ninh Binh |
Right conjunctiva |
11.0 |
|
8 |
Female |
50 |
Hung Yen |
Right conjunctiva |
10.0 |
|
9 |
Male |
50 |
Ha Nam |
Left conjunctiva |
12.5 |
|
10 |
Male |
36 |
Ha Nam |
Left subcutaneous side |
12.0 |
Table 2.Sequencing of the portion cox1 of different filarial species from GenBank compared with Dirofilaria repens* in Vietnam
Table 2.
|
Notation |
Origin |
Host |
Length |
Species |
GenBank |
Author |
|
GCA-VN1 |
Vietnam |
Human |
461 bp |
Dirofilaria repens*
|
- |
De, Le, and Chaia
|
|
GCH-VN2 |
Vietnam |
Human |
461 bp |
Dirofilaria repens*
|
- |
De, Le, and Chaia
|
|
GCD-VN3 |
Vietnam |
Human |
461 bp |
Dirofilaria repens*
|
- |
De, Le, and Chaia
|
|
Drep (ITA1) |
Italy |
- |
461 bp |
Dirofilaria repens
|
AJ271614 |
[8] |
|
Drep (ITA2) |
Italy |
- |
461 bp |
Dirofilaria repens
|
DQ358814 |
Serini et al. (GenBank) |
|
Dimm |
Australia |
Dog |
461 bp |
Dirofilaria immitis
|
AJ537512 |
[14] |
|
Bmal |
GenBank |
- |
461 bp |
Brugia malayi
|
AF538716 |
[15] |
|
Ovol |
GenBank |
- |
461 bp |
Onchocerca volvulus
|
AF015193 |
[16] |
Table 3.Percentage identity of nucleotide (above diagonal) and amino acid homology (below diagonal) of cox1 sequences of Vietnamese Dirofilaria repens and other Filaridae in GenBank
Table 3.
|
GCA- VN1 |
GCH- VN2 |
GCD- VN3 |
Drep (ITA1) |
Drep (ITA2) |
Dimm |
Bmal |
Ovol |
|
GCA-VN1 |
|
100 |
100 |
95 |
95 |
89 |
87 |
91 |
|
GCH-VN2 |
100 |
|
100 |
95 |
95 |
89 |
87 |
91 |
|
GCD-VN3 |
100 |
100 |
|
95 |
95 |
89 |
87 |
91 |
|
Drep (ITA1) |
100 |
100 |
100 |
|
99 |
90 |
87 |
92 |
|
Drep (ITA2) |
99 |
99 |
99 |
99 |
|
90 |
87 |
92 |
|
Dimm |
96 |
96 |
96 |
96 |
96 |
|
83 |
89 |
|
Bmal |
94 |
94 |
94 |
94 |
94 |
92 |
|
86 |
|
Ovol |
98 |
98 |
98 |
98 |
97 |
96 |
95 |
|