Abstract
There are 60 species of blood-feeding land leeches, 50 species belonging to the family Haemadipsidae and 10 species belonging to the family Xerobdellidae. Despite recent papers on the land leeches, their taxonomic identification is not fully understood, especially at a species level. In Korea, there have been no historical records of the terrestrial leeches, but recently an unrecorded blood-feeding land leech was discovered at Gageo-do (Island), Korea. Molecular analysis was used to identify the species of 29 leeches collected from Mt. Dock-Sil in Gageo-do. Conventional PCR was conducted using nuclear 18S rRNA and mitochondrial cytochrome c oxidase subunit 1 (CO1) genetic marker. The 18S rRNA sequences revealed that the leeches share 99.9% identity with Haemadipsa rjukjuana (inhabiting Taiwan), and the CO1 sequences revealed that the leeches are very close to H. rjukjuana (inhabiting Taiwan). The CO1 sequences were separated into 2 categories, 1 with 94.6% and the other with 94.3% similarity to the H. rjukjuana L00115A (inhabiting Taiwan). This new finding of the land leech is the first record in Korea. In addition, the north range of the distribution of the blood-feeding leech (Hirudiniformes: Haemadipisidae) should be reconsidered including Korea.
-
Key words: Haemadipsa rjukjuana, terrestrial leech, blood-feeding vector
INTRODUCTION
Leeches are segmented worms belonging to the phylum Annelida. Unlike other oligochaeta such as earthworms, they have 2 suckers. The majority of leeches inhabits in freshwater environments, but may also reside in terrestrial environments. They are generally classified into 2 groups: Rhyncobdellae (jawless leeches) and Arhynchobdellae (jawed leeches). Jawed leeches are known for blood-feeding habits and can be found in terrestrial and aquatic environments. Blanchard [
1] established Haemadipsidae to distinguish blood-feeding terrestrial leeches from their aquatic sanguivorous and carnivorous counterparts in Hirudininae [
2]. In addition, blood-feeding land leeches are separated into 2 families, Haemadipisidae and Xerobdellidae [
3] based on their phylogenetic analyses [
2,
4,
5,
6].
Land leeches belonging to the family Haemadipisidae are terrestrial hematophagous leeches. They exhibit high biodiversity especially in tropical regions with high humidity, such as the damp jungles and forests of South Asia, Southeast Asia, and Australia.
Haemadipsa, trignathous haemadipsid, is mostly abundant in Southeast Asia [
7]. Despite its high abundance throughout the range countries, the diversity and ecology of
Haemadipsa leeches have been poorly understood. Recent studies [
7,
8] have been conducted to identify
Haemadipsa leeches at a species level. Lai [
8] identified a new species using DNA barcoding which has been widely used [
9,
10,
11]. DNA barcoding system for animal species identification applies cytochrome
c oxidase subunit 1 (CO1) sequences as a genetic marker [
12]. Additionally, other studies analyzed distantly-related lineages of blood-feeding terrestrial leeches using nuclear 18S rRNA and 28S rRNA marker [
2].
The presence of
Torix tagoi Oka,
Hirudo nipponia Whitman 1886, and
Whitmania pigra (Murada, 1936) were first reported in Korea during the first half of the 20th century [
13]. In addition,
Glossiphonia weberi and
Hemiclepsis japonica were recorded [
14]. However, they described only freshwater leeches and did not apply molecular analysis. It has been known that some land leeches are living in islands of Korea, particularly in Gageo-do (Island), but yet to be documented scientifically. The goal of this study is to identify the taxonomic status of the land leeches newly found in Gageo-do based on molecular analysis. This is the first report on the presence of the terrestrial leech species in Korea.
MATERIALS AND METHODS
Study site and sample collection
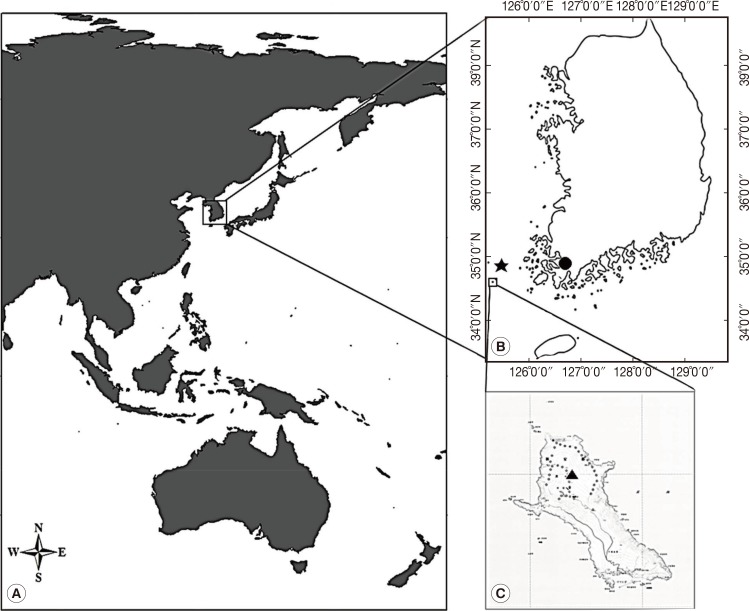
During July 25-28 of 2011, 40 leeches were collected on Mt. Dock-Sil (a.s.l. 639 m, E 125°07', N 34°04') on the Gageo-do, Shinan-gun (County), Jeollanam-do (Province), Korea (
Fig. 1). The island is located at the southwestern part of Mokpo City (136 km far from Mokpo), and its geographical coordinates is between N 34°06' and 35°02' and between E 125°05' and 125°09' (
Fig. 1). Leech collection was performed walking along the forest path to attract leeches, and then the leeches on legs or other parts were removed with tweezers as soon as possible (
Fig. 2A and B). The specimens were preserved in 70% alcohol for DNA extraction (
Fig. 2C and D). DNA extracted from 29 of 40 leeches were used for PCR.
The DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) was used for lysis of tissue and DNA extraction. The extracted DNA was stored at -20℃. PCR amplifications of 18S rRNA and COI gene fragments were carried out using the primers listed in
Table 1 [
15,
16]. The PCR amplification of 18S rRNA gene was performed by nested PCR. The initial PCR amplification was conducted using the primers A and B, yielding a 1.8 kb fragment. The subsequent PCR amplification was performed using 2 sets of nested primers A and Y or C and B to yield 2 short overlapping DNA fragments with approximately 1.2 kb in length each [
15]. All PCRs were conducted using the
HiPi PCR PreMix (Eplis-Biotech Inc., City, Country) in 20 µl total volume. Amplification reactions consisted of 1 unit/4 µl
HiPi Thermostable DNA polymerase in 250 mM Tris-HCl (pH 9.0), 80 mM (NH
4)
2SO
4, 10% DMSO, 8.75 mM MgCl
2, 0.05% bromophenol blue, 12% glycerol, stabilizer, 0.5 mM each primers, and 10-15 ng of DNA. All PCR reactions were performed in a PTC-200 thermal cycler (MJ Research Inc., Ramsey, Minnesota, USA). Amplified PCR products were separated by electrophoresis on 1.5% agarose gel and visualized with ethidium bromide under a SL-20 Image visualizer™ (Seolin Scientific Co., Seoul, Korea).
PCR amplicons were analyzed by direct sequencing. The GenBank accession numbers of 18S rRNA gene and COI gene sequences related to haemadipsoid leeches were listed in
Table 2. For 18S rRNA sequences, the 2 separated fragments (1.2 kb for each) were edited and reconciled using BioEdit version 7.0.9.0 [
17]. All sequences were manually edited at least 3 times. Alignment of sequences was carried out using MultiAlign [
18]. Consequently, both 1.7 kb of 18S rRNA and 649 bp of CO1 sequences were used for species identification and phylogenetic analysis. Analyses of the sequences were comparatively completed using the Blast Search option for 18S rRNA and CO1 sequences of leeches in GenBank database. The phylogenetic relationships between haplotypes of the leeches were reconstructed using the neighbor-joining (NJ) method [
19] under the Kimura 2-parameter (K-2-P) model [
20]. As references, corresponding sequences were selected from GenBank database (
Table 2). Also, corresponding sequences (n=8) were used to root the phylogenetic tree as outgroup (
Table 2). Confidence in estimated relationship was determined using the bootstrap approach [
21] obtained through 1,000 replicates with the same model as mentioned above. Both bootstrap analysis and phylogeny reconstruction were conducted using MEGA version 4.0 [
22].
RESULTS
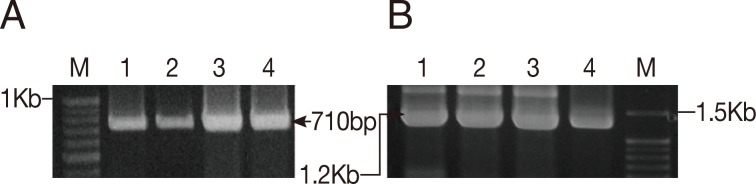
To determine the species identity and to evaluate the genetic relationship of the land leeches in Gageo-do, both 1.8 kb 18S rRNA and 710 bp CO1 gene sequences were amplified from leech samples (
Fig. 3). Of the 29 samples, 26 and 21 sequences were obtained for 18S rRNA and CO1 gene, respectively. The remaining samples were failed to yield clear sequences or proved to be other species, rather than
Haemadipsa spp. (data not shown).
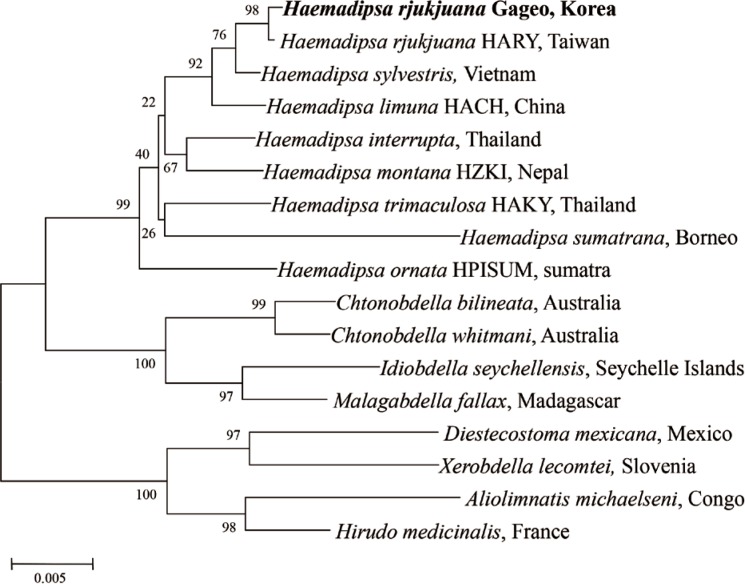
For determining the species identity of the Gageo-do land leeches, all sequences of the leeches were checked using the blast search from GenBank database. The results showed that 18S rRNA nucleotide sequences have 99.9% identity to the
H. rjukjuana isolate HARY (from Taiwan, HQ203097,
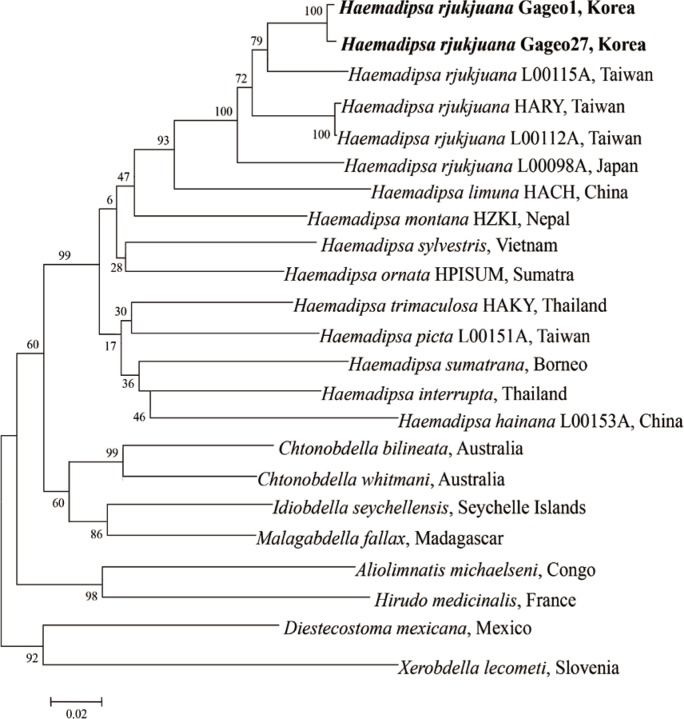
Fig. 4). They differed in only 2 base pairs of 1,706 bp. CO1 nucleotide also showed a similar pattern. The sequences were separated into 2 types,
H. rjukjuana CO1 type A and CO1 type B. Among 21 CO1 sequences that we acquired, type A sequences were 13 and type B sequences were 8. The type A and type B showed 94.6% (614/649 bp) and 94.3% (612/649 bp) similarity to the reference sequence of the
H. rjukjuana L00115A (from Taiwan, HQ322443,
Fig. 5), respectively.
For evaluating the genetic relationship of Gageo-do land leeches, both 18S rRNA and CO1 gene nucleotide sequences were independently analyzed with other reference sequences available in GenBank database. The NJ trees of terrestrial leech 18S rRNA and CO1 supported that
H. rjukjuana is monophyletic in a single clade with high bootstrap values (
Figs. 4,
5). For 18S rRNA gene (
Fig. 4), the genetic distance (
Ps) between
H. rjukjuana Gageo and
H. rjukjuana HARY was 0.1% (data now shown). In contrast, the
Ps between the
H. rjukjuana HARY and
H. sylvestris and between
H. rjukjuana Gageo and
H. sylvestris were 0.4 and 0.5%, respectively (data not shown). For CO1 gene (
Fig. 5), the
Ps betweeen the
H. rjukjuana L00115A and
H. limuma HACH is 14.6% (data now shown). However, the
Ps between the
H. rjukjuana L00115A and Gageo-do land leech 1 or 27 were 5.6% and 6.0%, respectively (data not shown). The phylogenetic tree also revealed that CO1 sequences of the Gageo-do land leeches had the closest relationship with
H. rjukjuana in Taiwan.
DISCUSSION
In general, leeches are divided into 2 groups, Rhynchobdellida Blanchard (jawless) and Arhynchobdellida Blanchard (jawed). Arhynchobdellid leeches have a muscular jaw for feeding and are classified into the Erpobdelliformes Caballero and the diverse Hirudiniformes Caballero [
23]. Hirudiniformes are subdivided into 5 families; Americobdellidae Canallero, Cylicobdellidae Ringuelet, Haemadipsidae Blanchard, Haemopidae Richardson, and Hirudinidae Whitman [
21,
24]. The remaining blood-feeding Hirudiniformes can be classified depending on their habitat preference as the semi-aquatic Hirudinidae and the terrestrial (or land) Haemadipsidae [
21]. Also, Saywer [
24] further consolidated Haemadipsinae with Haemadipisidae to create a single family, which is divided into the dougnathous (2-jawed) and trignathous (3-jawed) series, and included all land leech species. The trignathous leeches of the genus
Haemadipsa Tennent (1859) are widespread throughout the Indian subcontinent into southeast Asia and up to north-east Asia [
25].
Torix tagoi (Oka, 1925),
H. nipponia Whitman 1886,
W. pigra (Murada, 1936),
Glossiphonia werevi, and
Hemiclepsis japonica have been documented in Korea [
14]. However, these previous studies focused on morphological features of freshwater leeches but not terrestrial leeches [
26]. It has been previously known that terrestrial leeches inhabit the Gageo-do in Korea but none of researchers have done their taxonomy. This study documented the capture of terrestrial leeches on Mt. Dock-Sil of Gageo-do and their identification at a species level. Gageo-do is a west-southernmost island of Korea and located over the migratory routes of birds moving from Southeast Asia to Siberia. The total area and coastline length of Gageo-do is 9.18 km
2 and 22.0 km, respectively. The island which is composed of rocks and evergreen forests has a maritime climate. There is no weather station on Gageo-do, and according to the Heuksan-do weather station which is the nearest location from Gageo-do (70 km away), the mean temperature is 3.3-6.0℃ during winter and 22-27℃ during summer. The annual average temperature is 13.6℃, and the annual total precipitation is 1,126 millimeters. From May to August, there are lots of foggy weathers, and the fog tends to spread locally. Especially, Dock-Sil Mountain (a.s.l. 639 m) in Gageo-do is the highest mountain in Shinan-gun, and its internal forest generally shows a very humid condition throughout the year. Generally, terrestrial leeches are known to inhabit in tropical regions with high humidity, and in this regard the Mt. Dock-Sil serves as an ideal habitat for leeches. The terrestrial leeches on Mt. Dock-Sil possess an interior and a posterior sucker and move like extremely elastic inchworm. They suck the blood of human legs for about 30 min, after their blood-feeding, a bleeding scar is formed (
Fig. 2). They were regarded as the family Haemadipsidae but there were no preliminary data comparable to other land leeches.
As the result of 18S rRNA gene analysis of Gageo-do leeches (n=28/29 leeches), 26 sequences had 99.9% similarity to the
Haemadipsa rjukjuana isolate HARY (from Taiwan, HQ203097). The other 2 sequences were closer to the
Orobdella tsushimensis (from Japan, AB663653, data not shown). As the result of CO1 gene analysis (n=24/29 leeches), 21 were closest to the
H. rjukjuana. The 21 sequences were further divided into 2 types, Gageo 1 and Gageo 27, by only 1 bp. Both haplotypes had a 94.6% or 94.3% similarity to the
H. rjukjuana L00115A (from Taiwan, HQ322443). The remaining 3 sequences were similar to the
O. tsushimensis (Japan, AB679660, data not shown). For both genetic markers, the same result was obtained using CO1 gene. The phylogenetic trees also showed that Gageo-do leeches were closest to
H. rjukjuana (
Figs. 4,
5). It is unclear when the sanguivorous leeches start living on Gageo-do, but a possible hypothesis is that they were spread from other regions by humans (e.g., fisherman or tourists) or animals (e.g., migratory birds). However, further surveys are required to prove these hypotheses. In addition, according to the distribution map of Haemadipisidae (Borda and Siddall 2010), sub-distribution of the genus
Haemadipsa which have 3-jawed are widespread throughout India subcontinent into Southeast Asia and up to Northeast Asia. However, the boundary line of Northeast Asia is lined up to Japan. According to results of this study, the northern boundary line should be extended into Korea.
The main host of Haemadipsa land leeches are medium- or large-sized mammals primarily, including humans. We should try to identify animal species blood-sucked by leeches. Also, Haemadipsa land leeches should be confirmed whether they play as a vector or not for transmitting zoonotic pathogens.
The present study supports the presence of blood-feeding terrestrial leeches with a high genetic similarity to H. rjukjuana in Korea. This is the first record of sanguivorous land leeches (Hirudiniformes: Haemadipisidae) in Korea.
National Institute of Biological Resources, Korea1800-1844-301-260
Notes
-
We have no conflict of interest related to this study.
ACKNOWLEDGMENTS
This work was supported by the National Institute of Biological Resources (1800-1844-301-260), Korea.
References
- 1. Blanchard R. Hirudinees. Bollettino dei Musei Torino 1896;11:1-24.
- 2. Borda E, Oceguera-Figueroa A, Siddall ME. On the classification, evolution and biogeography of terrestrial haemadipsoid leeches (Hirudinida: Arhynchobdellida: Hirudiniformes). Mol Phylogenet Evol 2008;46:142-154.
- 3. Sket B, Trontelj P. Global diversity of leeches (Hirudinea) in freshwater. Hydrobiol 2008;595:129-137.
- 4. Trontelj P, Sket B, Steinbruck G. Molecular phylogeny of leeches: congruence of nuclear and mitochondrial rDNA data sets and the origin of bloodsucking. J Zool Syst Evol Res 1999;37:141-147.
- 5. Borda E, Siddall ME. Arhynchobdellida (Annelida: Oligochaeta: Hirudinida): phylogenetic relationships and evolution. Mol Phylogenet Evol 2004;30:213-225.
- 6. Kutschera U, Pfeiffer I, Ebermann E. The European land leech: biology and DNA-based taxonomy of a rare species that is threatened by climate warming. Naturwissenschaften 2007;94:967-974.
- 7. Ngamprasertwong T, Thirakhupt K, Panha S. Two new species of land leeches from Thailand (Hirudiniformes: Haemadipsidae). Nat Hist J Chulalongkorn Univ 2007;7:155-159.
- 8. Lai YT, Nakani T, Chen JH. Three species of land leeches from Taiwan, Haemadipsa rjukjuana comb.n., a new record for Haemadipsa pocta Moore, and an updated description of Tritetrabdella taiwana (Oka). Zookeys 2011;139:1-22.
- 9. Lai YT, Chang CH, Chen JH. Two new species of Helobdella Blanchard, 1896 (Hirudinida: Rhynchocdellida: Glossiphoniidae) from Taiwan, with a checklist of Hirudinea fauna of the Island. Zootaxa 2009;2068:27-46.
- 10. Siddall ME, Trontelj P, Utevsky SY, Nkamany M, Macdonald KS. Diverse molecular data demonstrate that commercially available medicinal leeches are not Hirudo medicinalis. Proc Biol Sci 2007;274:1481-1487.
- 11. Bely AE, Weisblat DA. Lessons from leeches: a call for DNA barcoding in the lab. Evol Dev 2006;8:491-501.
- 12. Hebert PD, Ratnasingham S, deWaard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci 2003;270(suppl 1):S96-S99.
- 13. Kang KW. Leech. Seoul, Republic of Korea. Academybook; 1995, p 90.
- 14. Paik EI, Park SR. A taxonomic study on two new records of freshwater leeches, Glossiphoniidae (Hirudinea: Rhynchobdellae) from Korea. Jayeongwahagyeongunonmunjip-Daeguhyoseonggatollikdaehakgyo 1992;44:497-502.
- 15. Apakupakul K, Siddall ME, Burreson EM. Higher level relationships of leeches (Annelida: Clitellata: Euhirudinea) based on morphology and gene sequences. Mol Phylogenet Evol 1999;12:350-359.
- 16. Folmer O, Back M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 1994;3:294-299.
- 17. Hall TA. BioEdit: a user-friendly biological sequence alignment deitor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999;41:95-98.
- 18. Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 1988;16:10881-10890.
- 19. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4:406-425.
- 20. Kimura MA. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980;16:111-120.
- 21. Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985;39:783-791.
- 22. Tamura K, Dundley J, Nei N, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 2007;24:1596-1599.
- 23. Borda E. Systematics and biodiversity of Arhynchobdellida (Oligochaeta: Hirudinida) with a focus on the evolutionary history of bloodfeeding terrestrial leeches. Michigan, USA. ProQuest. Information and Learning Company; 2007, p 223 UMI Microform 3288963.
- 24. Saywer RT. Leech Biology and Behavior: Volume II. Feeding, Biology, Ecology and Systematics. USA. Oxford Science Publications, Oxford University Press; 1986, p 360.
- 25. Borda E, Siddall ME. Insights into the evolutionary history of Indo-Pacific bloodfeeding terrestrial leeches (Hirudinida : Arhynchobdellida : Haemadipsae). Invertebr Syst 2010;24:456-472.
- 26. Park KH, Kim CH. Taxonomic studies on freshwater leeches (=Hirudinea) of Korea. Chungnam J Sci 1989;16:149-158.
Fig. 1Dock-Sil Mountain (▴, altitude 639 m) in Gageo-do (Island) (125°7"E, 34°4"N), Korea. Gageo-do is at Shinan-gun, Jeollanam-do (Province) and Korea's south-westernmost Island. Dock-Sil Mountain is the highest peak in Shinan-gun. (A) The geographical map of Asia. (B) The location of Mokpo (•), Heuksan-do (⋆), and Gageo-do, Korea. Gageo-do is 136 km distant from Mokpo and 70 km from Heuksan-do. (C) A map showing the topography of the Gageo-do and Dock-sil Mountain (▴).
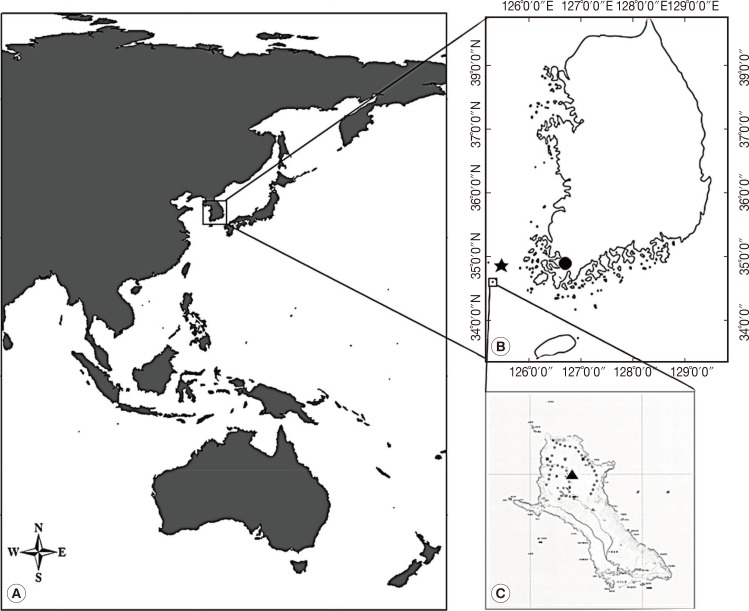
Fig. 2Terrestrial leeches collected from Gageo-do, Korea. (A) 10 min after feeding blood on a human leg. (B) 30 min after feeding on a human leg. (C, D) 70% alcohol-fixed terrestrial leeches after feeding.

Fig. 3Results of PCR amplification of COI and 18S rRNA genes from the Gageo-do land leech. (A) COI gene PCR amplicons (710 bp). (B) 18S rRNA gene amplicons (1.2 kb). Out of the total 1.8 kb 18S rRNA, PCR amplifications were conducted in 2 independent parts which are overlapped each other with approximately 1.2 kb in length; M, Elpis 100 bp marker; 1, 2, 3, and 4; Gageo land leeches sample.

Fig. 4A phylogenetic tree illustrating the genetic relationship of Haemadipsoid leeches using the neighbor-joining method implemented in Clustal X algorithm with Mega 4.0. 18S rRNA gene sequences of Haemadipsa rjukjuana obtained from Mt. Dock-Sil in Gageo-do, Shinan-gun, Jeollanam-do, Korea compared to previous reference sequences. Bold letters are sequences identified in this study.

Fig. 5Phylogenetic tree for COI gene showing the taxonomic status of haemadipsoid leeches using the neighbor-joining method implemented. COI gene sequences of Haemadipsa rjukjuana obtained from Mt. Dock-Sil in Gageo-do, Shinan-gun, Jeollanam-do, Korea. Bold letters are sequences identified in this study. H. rjukjuana Gageo 1 indicates COI type A and H. rjukjuana Gageo 27 indicates COI type B.
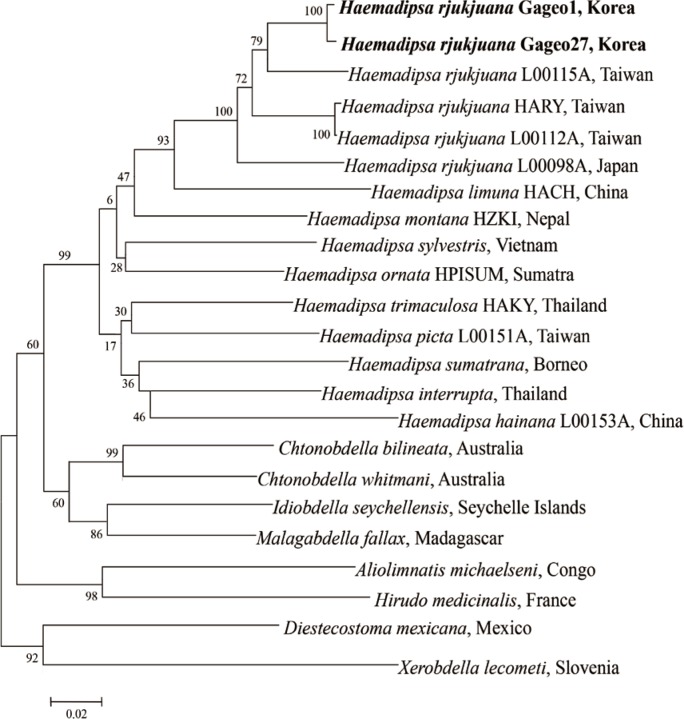
Table 1.Nucleotide sequences of PCR primers and conditions for amplification of 18S rRNA and COI genes
Table 1.
|
Target genes |
Name of PCR primers |
primer sequences (5´-3´) |
PCR product size |
References |
|
Nuclear 18S rRNAa |
A |
AACCTGGTTGATCCTGCCAGT |
1.2 kb |
Apakupakul et al. [15] |
|
Y |
CAGACAAATCGCTCC |
|
|
|
C |
CGGTAATTCCAGCTC |
1.2 kb |
|
|
B |
TGATCCTTCCGCAGGTTCACCT |
|
|
|
Mitochondrial CO1 |
LCO 1490 |
GGTCAACAAATCATAAAGATATTGG |
710 bp |
Folmer et al. [16] |
|
HCO 2198 |
TAAACTTCAGGGTGACCAAAAAATCA |
|
|
Table 2.Collection localities and GenBank accession numbers used for phylogenetic analyses of haemadipsoid leeches in this study
Table 2.
|
Taxon |
Locality |
GenBank accession no.
|
|
18S rNRA |
COI |
|
Haemadipsa rjukjuana Gageo (In this study) |
Gageo Islands, Korea |
KC524508a
|
- |
|
Haemadipsa rjukuana Gageo 1 (In this study) |
Gageo Islands, Korea |
- |
KC524509a
|
|
Haemadipsa rjukjuana Gageo 27 (In this study) |
Gageo Islands, Korea |
- |
KC524510a
|
|
Haemadipsa rjukjuana L00112A |
Taiwan |
- |
HQ322438 |
|
Haemadipsa rjukjuana L00115A |
Taiwan |
- |
HQ322443 |
|
Haemadipsa rjukjuana L00098A |
Ryukyu Islands, Japan |
- |
HQ322462 |
|
Haemadipsa hainana voucher L00153A |
Hainan Island, China |
- |
HQ322473 |
|
Haemadipsa picta voucher L00151A |
Taiwan |
- |
HQ322470 |
|
Haemadipsa rjukjuana isolate HARY |
Taiwan |
HQ203097 |
HQ203174 |
|
Haemadipsa limuna isolate HACH |
China |
HQ207191 |
HQ203169 |
|
Haemadipsa montana isolate HZKI |
Nepal |
HQ203105 |
HQ203182 |
|
Haemadipsa ornata isolate HPISUM |
Sumatra |
HQ203101 |
HQ203178 |
|
Haemadipsa trimaculosa isolate HAKY |
Thailand |
HQ203095 |
HQ203172 |
|
Haemadipsa interrupta
|
Thailand |
EU100069 |
EU100091 |
|
Haemadipsa sumatrana
|
Borneo |
AY425464 |
AY425446 |
|
Haemadipsa sylvestris
|
Vietnam |
AF116005 |
AF003266 |
|
Chtonobdella bilineata
|
Australia |
AF116006 |
AF003267 |
|
Chtonobdella whitmani
|
Australia |
EU100065 |
EU100087 |
|
Idiobdella seychellensis
|
Seychelle Islands |
EU100070 |
EU100094 |
|
Malagabdella fallax
|
Madagascar |
EU100071 |
EU100096 |
|
Diestecostoma mexicana
|
Mexico |
EU100068 |
EU100089 |
|
Xerobdella lecometi
|
Slovenia |
AF099947 |
EU100099 |
|
Aliolimnatis michaelseni
|
Congo |
AF116010 |
AF116029 |
|
Hirudo medicinalis
|
France |
AY786464 |
EU100093 |