Abstract
Echinococcus granulosus is the causative agent of cystic echinococcosis with medical and veterinary importance in China. Our main objective was to discuss the genotypes and genetic diversity of E. granulosus present in domestic animals and humans in western China. A total of 45 hydatid cyst samples were collected from sheep, humans, and a yak and subjected to an analysis of the sequences of mitochondrial cytochrome b (cytb) gene. The amplified PCR product for all samples was a 1,068 bp band. The phylogenetic analysis showed that all 45 samples were identified as E. granulosus (genotype G1). Ten haplotypes were detected among the samples, with the main haplotype being H1. The haplotype diversity was 0.626, while the nucleotide diversity was 0.001. These results suggested that genetic diversity was low among our samples collected from the west of China based on cytb gene analysis. These findings may provide more information on molecular characteristics of E. granulosus from this Chinese region.
-
Key words: Echinococcus granulosus, cytochrome b, sequence analysis, western China
Cystic echinococcosis (CE, also known as hydatid disease), a globally important and pathogenic zoonoses, is caused by the atiological agent,
Echinococcus granulosus. China is one of the most important endemic regions of CE [
1].
E. granulosus is now considered as a complex consisting of at least 4 species including
E. granulosus sensu stricto (G1-G3 genotypes),
E. equinus (G4), and
E. ortleppi (G5). There are controversies for considering G6 to G10 as 1 or 2 species, namely
E. canadensis and
E.intermedius [
2,
3,
4]. Among them,
E. granulosus s.s. is known to have a broad geographical distribution and a wide host range [
5]. G1-G3 genotypes are also named the sheep (G1), the Tasmanian sheep (G2), and the buffalo (G3) strains, respectively. In China, the sheep strain is the predominant epidemic strain, while G3 and G6 strains also exist [
6,
7,
8,
9,
10,
11,
12,
13,
14].
Increased knowledge of the genetic structure of a parasite could provide more information about its epidemiology and transmission, since genetic variants may affect infectivity and pathogenicity [
15]. In China, some mitochondrial and nuclear genes have been applied in the analysis of genetic information in
E. granulosus, including cytochrome
c oxidase subunit 1 (
cox1), NADH dehydrogenase 1 (
nad1), ATP synthase F0 subunit 6 (
atp6), cytochrome
b (
cytb), elongation factor-1 alpha (
ef1α), 12S, and 16S [
2,
13,
14,
16,
17]. However, the genetic diversity of
E. granulosus in China has never been characterized based on the complete
cytb gene, and only the partial
cytb gene have been reported [
17,
18]. Previous studies demonstrated that both the complete
cytb and
cox1 sequences showed 1 causative agent of CE in a cat from Russsia, i.e.,
E. granulosus s.s. (G1) [
19]. Therefore, it is feasible that we use the complete
cytb gene as a genetic marker to analyse genetic information of
E. granulosus. The main purpose of this study was to figure out the genotypes and genetic diversity of
E. granulosus present in western China.
A total of 45 hydatid cysts were obtained from several hosts (sheep, yak, and humans) in Tibet Autonomous Region, Qinghai, and Sichuan Province, China.
Table 1 summarizes the number and origin of isolates examined in the 3 localities. All samples were stored at -70℃. Genomic DNA was extracted from each cyst by proteinase K treatment and phenol-chloroform extraction. All DNA samples were stored at -20℃.
Based on the published complete mitochondrial genome of E. granulosus (GenBank accession no. AF297617), the cytb gene was amplified using the following primer pairs: forward 5'-AGATATTAGAGATGGAGGAG-3' and reverse 5'-ACCCGAGTTAATACAGTCAG-3'. The primers were designed by Primer 5.0 software, and the product was 1,609 bp that contained the complete sequence of cytb gene. PCR was performed in a 25 µl reaction mixture containing 1 µl of template (the genomic DNA of each sample was used as a template for PCR), 1 µl of each primer (10 pmol/µl), 12.5 µl of 2× Taq PCR MasterMix and 9.5 µl of ddH2O. Negative controls were always included in PCR reactions to assess possible contamination. The PCR protocol consisted of 30 thermal cycles with a denaturation step (90 sec at 94℃), a hybridization step (90 sec at 46℃) and an elongation step (90 sec at 72℃) for each cycle. Afterwards, 5 µl of the PCR product was run in a 1% agarose gel and stained with ethidium bromide to detect the PCR amplicons. Amplification products were purified by using the Universal DNA Purification Kit (Cowin Biotech, Beijing, China) and subjected to automated sequencing following subcloning into the pMD19-T vector (TaKaRa, Beijing, China). All amplicons were sequenced at least 3 times.
Multiple alignments of nucleotide sequences were performed using the Clustal X 1.83 software, with variation sites deleted from the alignments. Population diversity indices (nucleotide diversity, number of haplotypes, and haplotype diversity) were calculated using DnaSP 5.0 software [
20]. Phylogenetic trees were constructed from the alignments with the neighbour-joining method in MEGA 5.0 software, with
Echinococcus multilocularis as the outgroup. Confidence intervals were obtained via 1,000 bootstrap replications for each branch of the tree. The identification of haplotypes and construction of the haplotype network was undertaken by TCS 1.21 software using statistical parsimony [
21]. The network estimation was run at a 95% connection limit. Several sequences were obtained from GenBank for similarity comparison and phylogenetic analysis. The complete mitochondrial genome of
E. granulosus G1 (AF297617), G4 (AF346403), G5 (AB208063), G6 (AB235846) G7 (AB23587), and G8 genotype (AB235848) were served as reference sequences [
22,
23], and
E. multilocularis (AB018440) as the outgroup [
24]. Published mtDNA sequences of
cytb (AY278067 and AB622276) [
17,
19] were used for comparison.
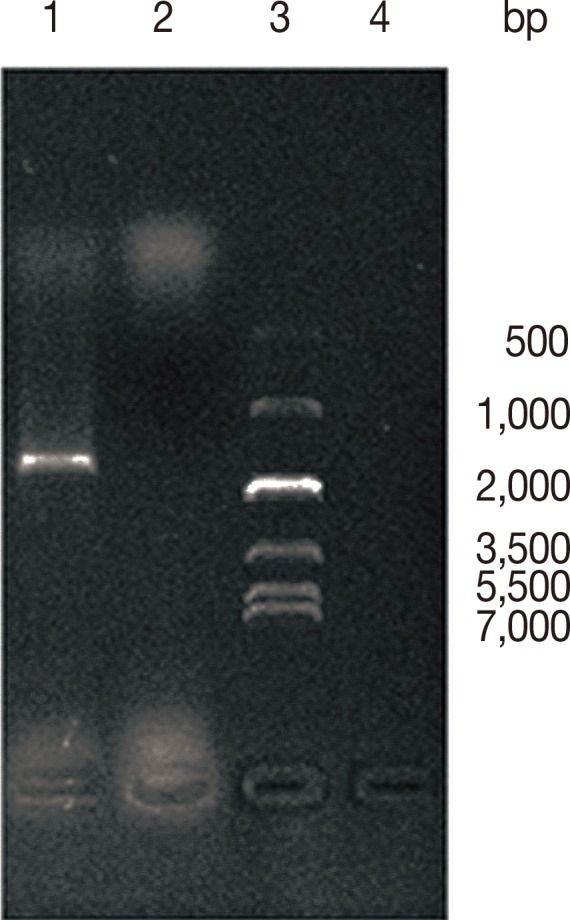
PCR amplification products of
cytb gene were analyzed by using 1% agarose gel electrophoresis, and the results showed that a 1,609 bp band was amplified between 1,000-2,000 bp, which was consistent with the expected length of the target fragment (
Fig. 1). DNA sequencing revealed that the
cytb gene sequences of all 45 samples contained 1,068 nucleotides, and sequences were deposited in GenBank with accession numbers KC709522 to KC709566. The average contents of A, T, C, and G were 19.4%, 47.7%, 8.9%, and 24.0%, respectively, revealing that AT content was greater than GC content. There were 11 mutation sites among these sequences of
cytb genes (
Table 2). Only nucleotide substitutions were detected, while deletions or insertion mutations were not observed. High similarity was shown among all the sequences obtained in this study (99.5% to 100%). In addition, all sequences in this study showed more than 88.0% identity with the previously published
cytb sequences for
E. granulosus genotypes.
We detected 10 haplotypes within the 45 isolates obtained from the west of China (
Table 2). It was indicated that each of the haplotypes possessed intermediate host specificity. Haplotype diversity (Hd) was 0.626, while nucleotide diversity (Pi) was 0.001. All the H1 haplotype sequences shared 100% identity with a partial
cytb gene sequence of a yak
E. granulosus strain from China (AY278067) [
17], and differed by only 1 nucleotide (a C/T substitution at the 263th site) from a cat
E. granulosus strain found in Japan (AB622276) [
19].
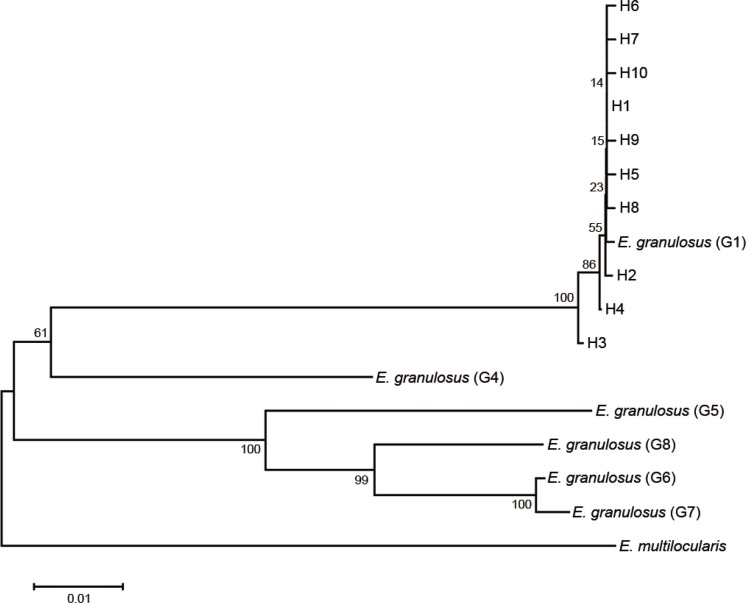
The phylogenetic tree showed that all isolates from the west of China were located on a single branch of the phylogenetic tree corresponding to
E. granulosus genotype G1, and all haplotypes identified in this study were distinguished from each other (
Fig. 2). The haplotype network of 45 isolates of
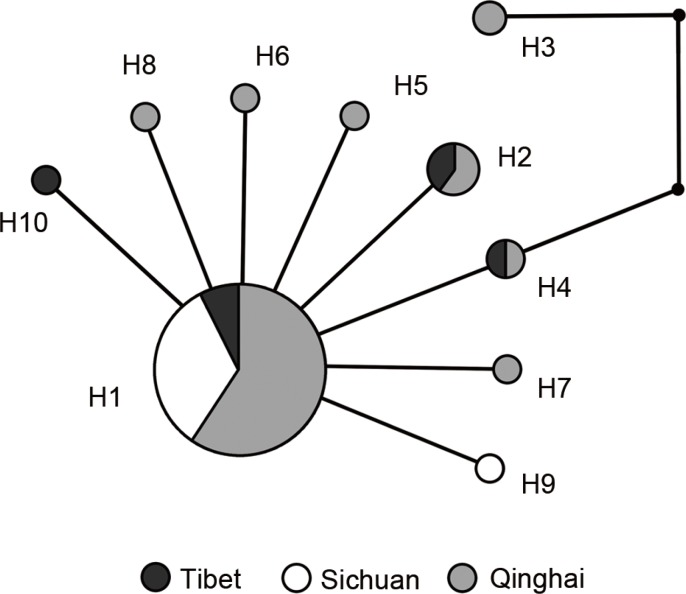
E. granulosus revealed that H1 was the predominant haplotype (
Fig. 3). H1 emitted 7 additional haplotypes and a branch containing 2 haplotypes (H3 and H4).
To date, a few studies have explored in detail genetic diversity of
E. granulosus based on
cytb gene. The partial nucleotide sequences of
cox1 (795 bp) and
cytb (568 bp) have amplified from a presumptive echinococcal liver lesion of a yak in Sichuan Province, and the results demonstrated that the yak was infected with
E. granulosus G1 [
17]. Based on partial sequences of
cytb gene (549 bp) and
rrnL (570 bp), Li et al. [
18] reported that 33 of 53 isolates obtained from humans in Sichuan and Qinghai were
E. granulosus G1, while the remaining 20 isolates were
E. multilocularis. In addition,
cytb (1,068 bp) and
cox1 (1,069 bp) gene sequences revealed that the causative agent of CE in a cat from Russia was
E. granulosus s.s. (G1) [
19]. In our study, using complete sequences of
cytb gene (1,068 bp) we identified all 45 isolates obtained from western China as belonging to the strain of
E. granulosus genotype G1. The data obtained in this study are more representative and comprehensive than the partial sequence. The complete sequence may provide a more effective evidence to verify the validity of the genotypes of
E. granulosus.
In recent years, studies based on nuclear and mitochondrial genes indicated that the G1 genotype is the predominant epidemic strain in the west of China, while the G3 and G6 strains also exist. Based on
cox1 (789 bp) and
ef1α (656 bp), the isolates of
E. granulosus s.s. (n = 181) and
E. canadensis G6 (n = 1) were identified successfully in Qinghai, Sichuan, and Xingjiang Province [
2]. Phylogenetic analysis based on
nad1 (332 bp) and
cox1 (450 bp) revealed that 28 isolates of
E. granulosus belonged to G1-G3 complex (
E. granulosus s.s.), and 2 isolates were placed in G6-G10 complex (
E. canadensis) in Qinghai [
11]. Yan et al. [
13] reported that 84 isolates of
E. granulosus from the Tibetan plateau in China were sequenced for the complete
nad1 (894 bp) and
atp6 (513 bp) gene, and the results showed that 82 isolates were identified as G1 genotype, while 2 as G3 genotype. Our findings are similar to previous results that G1 genotype of
E. granulosus is the main epidemic genotype in the west of China, but we did not detect G3 or G6 genotype in 45 samples of
E. granulosus.
Our results showed low genetic diversity among the 45 isolates of E. granulosus from the west of China. In addition, the similarities of cytb genes of 45 isolates was very high (99.5-100%), indicating that the genetic diversity among the 45 isolates was small.
In conclusion, G1 genotype was identified to be the only genotype in all 45 samples of E. granulosus from Tibet, Sichuan, and Qinghai provinces. The complete cytb gene investigated in this study enriched the genetic information among E. granulosus genotypes from the west of China.
National Key Technology R & D Program of China2006BAI06B09
Notes
-
We have no conflict of interest related to this study.
ACKNOWLEDGMENTS
The authors thank Zhongrong Jiang, Aiguo Yang, Shijin Deng, Li Guo, Dawa Tsering, Weishu Kong for their helpful assistance in collecting samples. This study was supported by grants from the National Key Technology R & D Program of China (no. 2006BAI06B09).
References
- 1. Thompson RC. The taxonomy, phylogeny and transmission of Echinococcus. Exp Parasitol 2008;119:439-446.
- 2. Nakao M, Yanagida T, Okamoto M, Knapp J, Nkouawa A, Sako Y, Ito A. State-of-the-art Echinococcus and Taenia: phylogenetic taxonomy of human-pathogenic tapeworms and its application to molecular diagnosis. Infect Genet Evol 2010;10:444-452.
- 3. Nakao M, Lavikainen A, Yanagida T, Ito A. Phylogenetic systematics of the genus Echinococcus (Cestoda: Taeniidae). Int J Parasitol 2013;43:1017-1029.
- 4. McManus DP. Current status of the genetics and molecular taxonomy of Echinococcus species. Parasitology 2013;140:1617-1623.
- 5. Schantz PM. Progress in diagnosis, treatment and elimination of echinococcosis and cysticercosis. Parasitol Int 2006;55(suppl):S7-S13.
- 6. Thompson RC, McManus DP. Aetiology: parasites and life-cyles. In Eckert J, Gemmell MA, Meslin FX, Pawlowski ZS eds, WHO/OIE Manual on Echinococcosis in Humans and Animals: a Public Health Problem of Global Concern. Paris, France. World Organisation for Animal Health; 2001, pp 1-19.
- 7. Pearson M, Le TH, Zhang LH, Blair D, Dai THN, McManus DP. Molecular taxonomy and strain analysis in Echinococcus. In Craig P, Pawlowski Z eds, Cestode zoononoses: Echinococcosis and Cysticercosis, an Emergent and Global Problem. Amsterdam, The Netherlands. IOS Press; 2002, pp 205-219.
- 8. Thompson RC, McManus DP. Towards a taxonomic revision of the genus Echinococcus. Trends Parasitol 2002;18:452-457.
- 9. Eckert J, Deplazes P. Biological, epidemiological, and clinical aspects of echinococcosis, a zoonosis of increasing concern. Clin Microbiol Rev 2004;17:107-135.
- 10. Bart JM, Abdukader M, Zhang YL, Lin RY, Wang YH, Nakao M, Ito A, Craig PS, Piarroux R, Vuitton DA, Wen H. Genotyping of human cystic echinococcosis in Xinjiang, PR China. Parasitology 2006;133:571-579.
- 11. Liu Q, Cao LL, Zhang YG, Xu D, Shang LM, Wang XL, Wei F, Xiao LH, Ma RL, Cai JS, Zhao QB. Genotypes of Echinococcus granulosus in animals from Yushu, northeastern China. Vector Borne Zoonotic Dis 2013;13:134-137.
- 12. Ma JY, Wang H, Lin GH, Craig PS, Ito A, Cai ZY, Zhang TZ, Han XM, Ma X, Zhang JX, Liu YF, Zhao YM, Wang YS. Molecular identification of Echinococcus species from eastern and southern Qinghai, China, based on the mitochondrial cox1 gene. Parasitol Res 2012;111:179-184.
- 13. Yan N, Nie HM, Jiang ZR, Yang AG, Deng SJ, Guo L, Yu H, Yan YB, Dawa T, Kong WS, Wang N, Wang JH, Xie Y, Fu Y, Yang DY, Wang SX, Gu XB, Peng XR, Yang GY. Genetic variability of Echinococcus granulosus from the Tibetan plateau inferred by mitochondrial DNA sequences. Vet Parasitol 2013;196:179-183.
- 14. Wang N, Wang JH, Hu DD, Zhong XQ, Jiang ZR, Yang AG, Deng SJ, Guo L, Dawa T, Wang SX, Gu XB, Peng XR, Yang GY. Genetic variability of Echinococcus granulosus based on the mitochondrial 16S ribosomal RNA gene. Mitochondrial DNA 2013;(published online: http://informahealthcare.com/doi/abs/10.3109/19401736.2013.840590)
- 15. Campbell G, Garcia HH, Nakao M, Ito A, Craig PS. Genetic variation in Taenia solium. Parasitol Int 2006;55(suppl):S121-S126.
- 16. Ma SM, Maillard H, Zhao HL, Huang X, Wang H, Geng PL, Bart JM, Piarroux R. Assessment of Echinococcus granulosus polymorphism in Qinghai Province, People's Republic of China. Parasitol Res 2008;102:1201-1206.
- 17. Xiao N, Qiu JM, Nakao M, Nakaya K, Yamasaki H, Sako Y, Mamuti W, Schantz PM, Craig PS, Ito A. Short report: identification of Echinococcus species from a yak in the Qinghai-Tibet Plateau region of China. Am J Trop Med Hyg 2003;69:445-446.
- 18. Li TY, Ito A, Nakaya K, Qiu J, Nakao M, Zhen R, Chen X, Giraudoux P, Craig PS. Species identification of human echinococcosis using histopathology and genotyping in northwestern China. Trans R Soc Trop Med Hyg 2008;102:585-590.
- 19. Konyaev SV, Yanagida T, Ivanov MV, Ruppel VV, Sako Y, Nakao M, Ito A. The first report on cystic echinococcosis in a cat caused by Echinococcus granulosus sensu stricto (G1). J Helminthol 2012;86:391-394.
- 20. Rozas J, Sanchez-Delbarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 2003;19:2496-2497.
- 21. Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol 2000;9:1657-1659.
- 22. Le TH, Pearson MS, Blair D, Dai N, Zhang LH, McManus DP. Complete mitochondrial genomes confirm the distinctiveness of the horse-dog and sheep-dog strains of Echinococcus granulosus. Parasitology 2002;124:97-112.
- 23. Nakao M, McManus DP, Schantz PM, Craig PS, Ito A. A molecular phylogeny of the genus Echinococcus inferred from complete mitochondrial genomes. Parasitology 2007;134:713-722.
- 24. Nakao M, Okamoto M, Sako Y, Yamasaki H, Nakaya K, Ito A. A phylogenetic hypothesis for the distribution of two genotypes of the pig tapeworm Taenia solium worldwide. Parasitology 2002;124:657-662.
Fig. 1Electrophoresis results of amplification product of cytb gene. Lane 1, E. granulosus DNA; Lane 2, template free; Lane 3, marker; Lane 4, primers free.

Fig. 2Neighbour-Joining phylogenetic tree of complete mitochondrial Echinococcus granulosus cytb sequences showing the phylogenetic relationship of haplotypes, H1 to H10.

Fig. 3Haplotype network for the 10 haplotypes (H1 to H10) identified among 45 isolates of Echinococcus granulosus collected from western China.

Table 1.Geographic origin and host of China isolates of Echinococcus granulosus identified by DNA sequencing of mitochondrial cytb gene
Table 1.
|
Province |
Host
|
Total |
|
Sheep |
Human |
Yak |
|
Qinghai |
28 |
0 |
0 |
28 |
|
Sichuan |
0 |
9 |
1 |
10 |
|
Tibet |
7 |
0 |
0 |
7 |
|
Total |
35 |
9 |
1 |
45 |
Table 2.Variation sites of cytb gene of Echinococcus granulosus from the west of China
Table 2.
|
Haplotype |
Locality |
Host |
Na
|
97 |
120 |
180 |
285 |
471 |
680 |
768 |
850 |
888 |
984 |
1,038 |
|
H1 |
Qinghai, Sichuan, and Tibet |
Sheep Human |
27 |
A |
A |
A |
T |
T |
C |
T |
C |
G |
C |
G |
|
H2 |
Qinghai, Tibet |
Sheep |
5 |
- |
G |
- |
- |
- |
- |
- |
- |
- |
- |
- |
|
H3 |
Qinghai |
Sheep |
3 |
G |
- |
G |
- |
- |
- |
- |
T |
- |
T |
- |
|
H4 |
Qinghai, Tibet |
Sheep |
4 |
- |
- |
G |
- |
- |
- |
- |
- |
- |
- |
- |
|
H5 |
Qinghai |
Sheep |
1 |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
A |
|
H6 |
Qinghai |
Sheep |
1 |
- |
- |
- |
- |
C |
- |
- |
- |
- |
- |
- |
|
H7 |
Qinghai |
Sheep |
1 |
- |
- |
- |
C |
- |
- |
- |
- |
- |
- |
- |
|
H8 |
Qinghai |
Sheep |
1 |
- |
- |
- |
- |
- |
- |
- |
- |
A |
- |
- |
|
H9 |
Sichuan |
Human |
1 |
- |
- |
- |
- |
- |
T |
- |
- |
- |
- |
- |
|
H10 |
Tibet |
Sheep |
1 |
- |
- |
- |
- |
- |
- |
C |
- |
- |
- |
- |