Abstract
The rumen parasite, Gastrothylax crumenifer (Platyhelminthes: Gastrothylacidae), is a highly pathogenic trematode parasite of goat (Capra hircus). It sucks blood that causes acute disease like anemia, and severe economic losses occur due to morbidity and mortality of the ruminant infected by these worms. The study of these rumen paramphistomes, their infection, and public health importance remains unclear in India especially in the western part of state Uttar Pradesh (U.P.), Meerut, India, where the goat meat consumption is very high. This paper provides the molecular characterization of G. crumenifer recovered from the rumen of Capra hircus from Meerut, U.P., India by the partial sequence of 28S rDNA. Nucleotide sequence similarity searching on BLAST of 28S rDNA from parasites showed the highest identity with those of G. crumenifer from the same host Capra hircus. This is the first report of molecular identification of G. crumenifer from this part of India.
-
Key words: Gastrothylax crumenifer, goat (Capra hircus), ribosomal DNA, Uttar Pradesh, India
The rumen worm,
Gastrothylax crumenifer [
1,
2], is a blood-sucking trematode parasite found in the rumen of
Capra hircus,
Bos indicus,
Bubalus bubalis, and
Ovis aries. The infection causes anemia and accidental death of the animals which is serious and a major animal health problem [
3]. The prevalence of this parasite is very high in tropical countries [
4,
5]. Some reports specified that, in tropical and subtropical areas,
Paramphistomum cervi,
P. Epiclitum, and
G. crumenifer are the most prevalent parasites which cause the disease in ruminants [
6,
7,
8].
Goats are a treasure home for different helminth parasites, thus, it is difficult to discriminate them because of their morphological similarity. Identification of parasites is usually done by microscopic observations of parasite's characters like the acetabulum, pharynx, and terminal genitalium. For any parasitic infection, accurate identification of the species involved is the first vital step for subsequent studies in the field of epidemiology, physiology, and immunology of the parasite. Therefore, it is important to identify the species found in goat on the basis of molecular characterization. Since
G. crumenifer has been regarded as a valid species of the genus
Gastrothylax on the basis of morphological characters [
9]. It has been recorded from various localities in India, such as Uttar Pradesh, Jammu and Kashmir, Meghalaya, Himachal Pradesh, and Rajasthan [
10,
11,
12,
13,
14], but no previous molecular studies have been done in Meerut region. Paramphistomiasis is important diseases of ruminants despite that little attention have received with respect to use of molecular tools for their validation, particularly in Indian region.
In the present work, we report molecular identification of
G. crumenifer on the basis of sequences of the large subunit (LSU) ribosomal RNA (rRNA) 28S gene by using the PCR amplification and sequence analysis. The LSU or 28S rRNA is often regarded as a good phylogenetic marker, and its usage has provided a more fruitful resolution among the Metazoa [
15]. It has been widely used to rectify species phylogenies of digenetic trematodes [
16,
17,
18].
Flukes were collected from the rumens of freshly slaughtered goats at abattoirs located in the Sotiganj, Meerut (29°01' N, 77°45' E) of state U.P., western India. The flukes were washed extensively in saline solution, and some were stored at -20℃ until DNA extraction and molecular analysis were carried out. Others were used for morphological identification by optical and scanning electron microscopy (SEM). For morphological studies, the parasites were fixed in 70% ethanol for 24 hr and washed several times with water. They were then stained with Borax carmine, dehydrated through increasing grades of ethanol, and mounted in Canada balsam. The slides have been deposited in the Museum, Department of Zoology, Chaudhary Charan Singh University, U.P., India, under the voucher no. HS.TR/2013/01. For SEM, the specimens were fixed overnight with 3.5% glutaraldehyde. After then, the specimens were dehydrated through graded alcohol series and dried, mounted on aluminum stubs, and sputter coated with gold for 1 min. The observation and image acquisition was made in a JEOL-NeoScope JCM-5000 (NIKON-Type 108) SEM at 10 kV, according to the manufacturer's instructions.
Three flukes from each 2 ruminant in the study were used for extraction of genomic DNA. The genomic DNA extraction was performed on individual flukes using the Qiagen Tissue kit (QIAGEN, Hilden, Germany) according to the instructions specified by the manufacturer and stored at -20℃. The 28S sequences was amplified by PCR using the primers 28S F (5'-TAGGTCGACCCGCTGAAYTTAAGCA-3') as described by Littlewood et al. [
19] and 28S R (5'-GCTATCCTGAGGGAAACTTCG-3') as described by Pawlowski et al. [
20] with the annealing temperature 57℃. The PCR cycle, product purification, and sequencing reaction were performed as described by Chaudhary and Singh [
21] with the same primers used in this work. Sequences of 28S were aligned using the programmes Clustal W and MUSCLE (3.7) for phylogenetic analysis. Pairwise distances were calculated using the Tamura-Nei substitution model using the software MEGA 6 [
22]. Subsequent phylogenetic analysis of rDNA sequences was performed using the PhyML method with bootstrap values for n=100 replicates [
23]. The obtained nucleotide sequence is available in GenBank, NCBI under accession no. KJ559529.
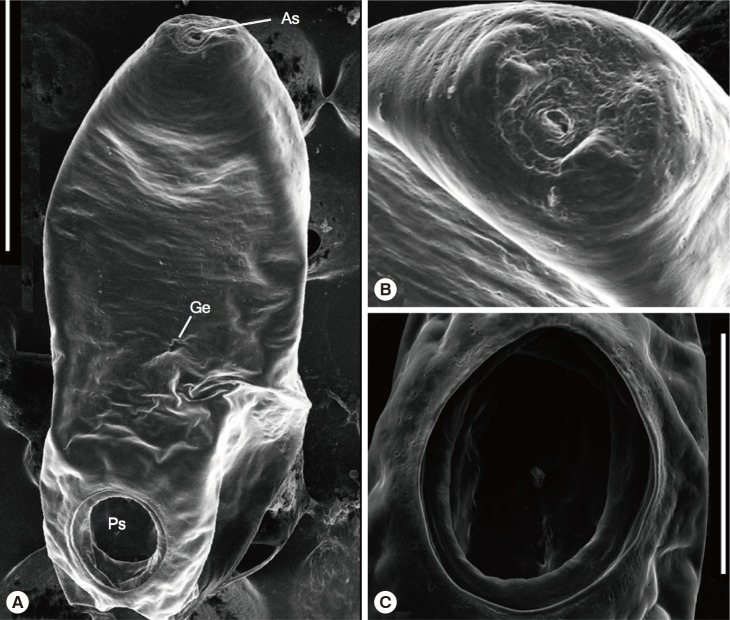
The stained slides of the parasite were examined using light microscopy. The SEM images show the full view of the worm (
Fig. 1), with its oral and posterior suckers (acetabulum). The sequences from each DNA sample were identical without any ambiguities. The complete sequence of the 28S from
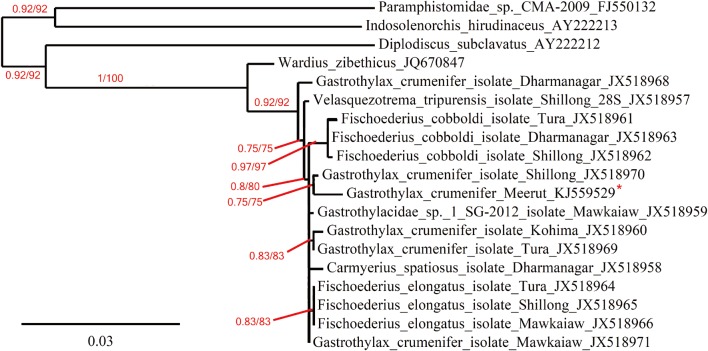
G. crumenifer was 1,293 bp long. The nucleotide sequence similarity search for 28S rDNA from the parasites of Meerut region showed 98% identity with the same species (JX518970) in GenBank collected from the same host from northern part of India (
Fig. 2). Beside the host
C. hircus, from India,
G. crumenifer was also reported from
B. indicus,
B. Frontalis, and
B. bubalis. We also compared our sequences with the sequence data of
G. crumenifer (JX518968, JX518960, JX518971, JX518969) obtained from these hosts to reveal the molecular characteristics (
Fig. 2). Based on phylogenetic analyses,
G. crumenifer was grouped with the same species isolated from
C. hircus (JX 518970) (
Fig. 2), both isolates were genetically close to each other. Maximum likelihood and Bayesian inference both placed
G. crumenifer (KJ559529 and JX518970) in a same clade (
Fig. 2) and gave highly similar tree topology for the position of
G. crumenifer (
Fig. 2). The Gastrothylacidae species forming a well-supported clade (100%) with 1 species of the Paramphistomatidae (JQ670847) (
Fig. 2).
Among ruminants, of the different paramphistomoid families, various species of parasites mainly the members of Paramphistomidae and Gastrothylacidae, cause a disease collectively known as paramphistomiasis [
24]. The family Gastrothylacidae is composed of 4 genera viz.,
Carmyerius Stiles and Goldberger, 1910 [
25],
Fischoederius Stiles and Goldberger, 1910 [
25],
Gastrothylax Poirier, 1883 [
2], and
Velasquezotrema Eduardo and Javellana, 1987 [
26]. Due to thick bodies, it is hard to see the internal parts of these worms; that is why it is difficult to identify these worms. Morphological differences based on stained and mounted specimens have been widely used for discrimination of platyhelminthes species [
27], but it is not possible to distinguish them on the basis of clinical, pathological, or immunological findings as they are morphologically very similar [
28]. Therefore, in order to gain better understanding of the phylogeny of this group of parasites needs the use of molecular tools.
The results of the present study confirmed that 28S ribosomal DNA region is a useful molecular marker for species identification and platyhelminthes phylogeny [
29,
30,
31,
32]. Phylogenetic analyses results based on 28S sequences revealed that
G. crumenifer from Meerut is closely related with the
G. crumenifer obtained from Shillong, and
G. crumenifer from infected goats at Meerut region showed to be identical to other
G. crumenifer corresponding sequences in GenBank, in accordance with previous studies [
32,
33]. It is also confirmed that there is a highly conserved character of the 28S gene among
G. crumenifer isolates from different regions of India [
32,
33]. The closeness of
G. crumenifer from Meerut and Shillong regions is not surprising as they are the same species from 2 distantly apart locations in India (western India and northern India) and depicted within the same clade. The results also corroborate that the
G. crumenifer species prevalent in India have not been identified properly due to the lack of genetic data. To determine the correct phylogeny and identification between the related species to this genus, further studies with additional molecular markers are needed.
This study reports the first molecular diagnosis and characterization of G. crumenifer from Meerut region, U.P., India. The current study is the first step toward expanding our knowledge of paramphistome fauna in this area. However, further studies are required to detect other paramphistome species, which may be concurrently infecting the same host or different hosts. Additional epidemiological and experimental studies must be undertaken to obtain specimens from other host samples from Meerut region, and further researches are required to determine the presence or absence of other species of paramphistomes in this region.
University Grants Commission (UGC), New Delhi, IndiaF1-17.1/2013-14/RGNF-2013-14-SC-UTT-48647
Notes
-
Authors declare that they have no conflict of interests.
ACKNOWLEDGMENTS
We wish to acknowledge the support of the Head, Department of Zoology, Chaudhary Charan Singh University, Meerut (U.P.), India, for assistance. This work was funded by a grant from the Junior Research Fellowship (JRF) Grant under Rajiv Gandhi National Fellowship (RGNF) (grant no. F1-17.1/2013-14/RGNF-2013-14-SC-UTT-48647/(SA-3rd/website) to AK by University Grants Commission (UGC), New Delhi, India.
References
- 1. Creplin F. Beschreibung zweier neuen Amphistomen-Artenaus dem zebu-Ochsen. Arch Naturgesch 1847;13:30-35.
- 2. Poirier MJ. Description d'helminthes nouveaux du Palonia frontalis. Bull Soc Philomat Paris 1883;7:73-80.
- 3. Hanna REB, Williamson DS, Mattison RG, Nizami WA. Seasonal reproduction in Paramphistomum epiclitum and Gastrothylax crumenifer, rumen paramphistomes of the Indian water buffalo and comparison with the biliary paramphistome Gigantocotyle explanatum. Int J Parasitol 1988;18:513-521.
- 4. Rangel-Ruiz LJ, Albores-Brahms ST, Gamboa-Aguilar J. Seasonal trends of Paramphistomum cervi in Tabasco, Mexico. Vet Parasitol 2003;116:217-222.
- 5. Ahmad T, Reshi ML, Chesti MZ, Tanveer S, Shah ZA. A case report of Gastrothylax crumenifer incidence in sheep in Kashmir Valley. Iraq J Vet Sci 2013;27:71-72.
- 6. Gupta PP, Singh B, Dutt SC. Note on amphistomiasis in an adult buffalo. Indian Vet J 1978;55:491-492.
- 7. Wang CR, Qiu JH, Zhu XQ, Han XH, Ni HB, Zhao JP, Zhou QM, Zhang HW, Lun ZR. Survey of helminths in adult sheep in Heilongjiang Province, People's Republic of China. Vet Parasitol 2006;140:378-382.
- 8. Panyarachun B, Ngamniyom A, Sobhon P, Anuracpreeda P. Morphology and histology of the adult Paramphistomum gracile Fischoeder, 1901. J Vet Sci 2013;14:425-432.
- 9. Sey O. Revision of the family Gastrothylacidae Stiles et Goldberger, 1910 (Trematoda: Paramphistomata). Acta Zool Acad Sci Hung 1983;29:223-252.
- 10. Srivastava HD. A survey of the incidence of helminth infection in India at the Imperial Veterinary Research Institute, Izatnagar. Indian J Vet Sci Anim Husb 1946;15:146-148.
- 11. Bali HS. A survey of helminth parasites of sheep (Ovis aries) in Jammu and Kashmir. J Anim Health Product 1976;6:25-32.
- 12. Soota TD, Ghosh RK. On some trematodes from Meghalaya. Rec Zool Surv India 1977;73:111-122.
- 13. Tandon V, Sharma V. Amphistome fauna of ruminants in Himachal Pradesh. Indian J Parasitol 1981;5:241-245.
- 14. Swarnakar G, Roat K, Sanger B, Kumawat A. Anthelminthic effect of Trigonella foenum-graecum on tegument of Gastrothylax crumenifer in cattle of Udaipur, India. Int J Curr Microbiol App Sci 2014;3:599-606.
- 15. Medina M, Collins AG, Silberman JD, Sogin ML. Evaluating hypotheses of basal animal phylogeny using complete sequences of large and small subunit rRNA. Proc Nat Acad Sci USA 2001;98:9707-9712.
- 16. Snyder SD, Tkach VV. Phylogenetic and biogeographical relationships among some holarctic frog lung flukes (Digenea: Haematoloechidae). J Parasitol 2001;87:1433-1440.
- 17. Tkach VV, Pawlowski J, Mariaux J, Swiderski Z. Molecular phylogeny of the suborder Plagiorchiata and its position in the system of Digenea. In Littlewood DTJ, Bray RA eds, Interrelationships of the Platyhelminthes. London, UK. Taylor and Francis; 2001, pp 186-193.
- 18. León-Règagnon V, Paredes-Calderón EL. Haematoloechus danbrooksi n. sp (Digenea: Plagiorchioidea) from Rana vaillanti from Los Tuxtlas, Veracruz, Mexico. J Parasitol 2002;88:1215-1221.
- 19. Littlewood DT, Curini-Galletti M, Herniou EA. The interrelationships of Proseriata (Platyhelminthes: Seriata) tested with molecules and morphology. Mol Phylogenet Evol 2000;16:449-466.
- 20. Pawlowski J, Bolivar I, Guiard-Maffia J, Gouy M. Phylogenetic position of foraminifera inferred from LSU rRNA gene sequences. Mol Biol Evol 1994;11:929-938.
- 21. Chaudhary A, Singh HS. Phylogenetic study of nine species of freshwater monogeneans using secondary structure and motif prediction from India. Bioinformation 2012;8:862-869.
- 22. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 2013;30:2725-2729.
- 23. Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 2008;36:W465-W469.
- 24. Sanabria REF, Romero JR. Review and update of paramphistomosis. Helminthologia 2008;45:64-68.
- 25. Stiles CW, Goldberger J. A study of the anatomy of Watsonius (n.g.) watsoni of man and of nineteen allied species of mammalian trematode worms of the superfamily Paramphistomoidea. Bull Hyg Lab (Public Health and Marine-Hospital Service of the United States). 1910, (60):p 259.
- 26. Eduardo SL, Javellana CRH. A new genus, Velasquezotrema, for Fischoederius brevisaccus Eduardo, 1981. Philip J Vet Med 1987;24:29-34.
- 27. Miyazaki I. Lung flukes in the world. Morphology and life history. In Sasa M ed, A symposium on epidemiology of parasitic diseases. Tokyo, Japan. International Medicine Foundation of Japan; 1974, pp 101-135.
- 28. Lotfy WM, Hillyer GV. Fasciola species in Egypt. Exp Pathol Parasitol 2003;11:9-22.
- 29. Jovelin R, Justine JL. Phylogenetic relationships within the polyopisthocotylean monogeneans (Platyhelminthes) inferred from partial 28S rDNA sequences. Int J Parasitol 2001;31:393-401.
- 30. Choudhury A, Rosas Valdez R, Johnson RC, Hoffmann B, Pérez-Ponce de Leon G. The phylogenetic position of Allocreadiidae (Trematoda: Digenea) from partial sequences of the 18S and 28S ribosomal RNA genes. J Parasitol 2007;93:192-196.
- 31. Atopkin DM. Genetic characterization of the Psilotrema (Digenea: Psilostomatidae) genus by partial 28S ribosomal DNA sequences. Parasitol Int 2011;60:541-543.
- 32. Shylla JA, Ghatani S, Tandon V. Utility of divergent domains of 28S ribosomal RNA in species discrimination of paramphistomes (Trematoda: Digenea: Paramphistomoidea). Parasitol Res 2013;112:4239-4253.
- 33. Ghatani S, Shylla JA, Tandon V, Chatterjee A, Roy B. Molecular characterization of pouched amphistome parasites (Trematoda: Gastrothylacidae) using ribosomal ITS2 sequence and secondary structures. J Helminthol 2012;86:117-124.
Fig. 1(A) Whole-body topography of Gastrothylax crumenifer (Creplin, 1847) Poirier, 1883. (B) An enlarged view of the anterior sucker. (C) An enlarged view of the posterior sucker (acetabulum). Scale bars=500 µm (A), 100 µm (B, C). As, anterior sucker; Ge, genital canal; Ps, posterior sucker.

Fig. 2Phylogenetic tree based on 28S sequence data and the numbers represent Bayesian probabilities/bootstap values from 100 replicates (maximum likelihood). Bootstrap values of <50% are not shown. Asteriks shows the species sequenced in this study.