Abstract
Introduction of double-stranded RNA (dsRNA) into some cells or organisms results in degradation of its homologous mRNA, a process called RNA interference (RNAi). The dsRNAs are processed into short interfering RNAs (siRNAs) that subsequently bind to the RNA-induced silencing complex (RISC), causing degradation of target mRNAs. Because of this sequence-specific ability to silence target genes, RNAi has been extensively used to study gene functions and has the potential to control disease pathogens or vectors. With this promise of RNAi to control pathogens and vectors, this paper reviews the current status of RNAi in protozoans, animal parasitic helminths and disease-transmitting vectors, such as insects. Many pathogens and vectors cause severe parasitic diseases in tropical regions and it is difficult to control once the host has been invaded. Intracellularly, RNAi can be highly effective in impeding parasitic development and proliferation within the host. To fully realize its potential as a means to control tropical diseases, appropriate delivery methods for RNAi should be developed, and possible off-target effects should be minimized for specific gene suppression. RNAi can also be utilized to reduce vector competence to interfere with disease transmission, as genes critical for pathogenesis of tropical diseases are knockdowned via RNAi.
-
Key words: RNAi, Parasitic protozoa, Helminth, Insect vector
INTRODUCTION
Genomes of hundreds of organisms have been sequenced [
1], holding the potential to reveal gene functions in those organisms. Genome sequences will enable researchers to develop more comprehensive approaches to understanding the biology of living organisms [
2]. However, functions of over half of the genes in sequenced species remain unknown, needing annotation to fully appreciate the biological functions of those genes. RNA interference (RNA
i) has been widely used as a molecular tool by which target transcripts can be sliced, resulting in the reduction of mRNAs for protein expression [
3]. Therefore, phenocopies of loss of function mutations are possible by RNA
i gene targeting.
While other genetic modifications such as germline transformation alter the genotypes of target organisms, introduction of double-stranded RNA (dsRNA) changes only the phenotypes of organisms without altering genotypes. dsRNA reduces the transcripts of specific mRNA, instead of deleting or inserting a gene like genetic modifications [
4]. Since its discovery in petunias, RNA
i has been used to suppress or silence specific mRNA transcripts in many different organisms [
5-
14]. The RNA
i technique has been frequently used for analyzing gene function and has the potential to be a tool for disease therapy by interfering with vector competence or pathogen development [
15].
RNA
i appears promising in silencing gene expression in parasitic pathogens such as protozoans and helminths, as well as disease vectors by specific target mRNA interference [
15]. Analysis of gene functions in pathogens of infectious disease and their vectors is important for research in drug development, and the silencing effects may be directly employed to control parasite transmission and development.
However, there are some practical obstacles when using RNA
i in tropical disease pathogens. RNA
i machinery may have been eliminated in certain parasitic protozoans, so the presence of a RNA
i pathway should be confirmed before utilization [
16]. An ideal delivery method should be established to generate optimal RNA
i effects. dsRNA-generating vector systems may be employed to solve the issue of transient RNA
i effects. Finally, off-target effects such as non-specific lethality or non-specific gene silencing should be avoided in order to maximize the effect of specific gene silencing.
This review focuses on the use of RNAi in protozoans, parasitic helminths and insect vectors that cause tropical diseases. In addition, RNAi pathways in insect vectors are reviewed in connection with other model insect species such as Drosophila melanogaster and the silkworm because insects share many common RNAi molecules. Moreover, well established RNAi delivery methods and protocols are also highly effective among other insect species including both vector and non-vector insects. Thus, it is appropriate here to review RNAi pathways and biological roles in non-vector species as they can provide insight into the biology and applications of RNAi in vector mosquitoes of diseases of medical importance.
DISCOVERY OF RNAi
RNA
i has been a new research tool to study genetic functions and therapeutics since the suppression of mRNA transcripts by dsRNA was discovered [
8]. RNA interference is present in various organisms including plants,
Caenorhabditis elegans [
7], adult mice [
10], chicken embryos [
17], trypanosomes [
6,
11], fungi [
18], as well as various insect species.
The phenomenon of gene silencing by RNA
i was first discovered in petunias [
4,
14]. It was shown that over-expression of the transgenic chalcone enzyme did not increase flower coloration as expected. The chalcone enzyme is responsible for flower coloration, but the transgenic flowers lost their pigmentation partially or completely. This phenomenon was called co-suppression since the mRNA of the transgene and an endogenous chalcone enzyme gene were suppressed at the same time; however, it was not known what factors caused the co-suppression.
Although dsRNAs were proven to be RNA
i inducing factors, it was initially speculated that foreign antisense RNAs hybridized with target mRNAs, causing the reduction in target gene expression [
19]. This hypothesis seemed feasible until Guo and Kemphues [
8] found a perplexing result; not only could antisense RNA of the
par-1 gene reduce expression in the nematode
C. elegans, but also sense RNA of
par-1 could suppress its endogenous target gene. This phenomenon was further investigated by Fire and Mello who clearly demonstrated that introduction of dsRNAs of a target gene induced gene silencing [
7]. They showed dsRNA was more effective than either sense or antisense RNA alone in producing an abnormal
unc-22 phenotype, which impairs motility in
C. elegans. This was the first unequivocal demonstration that dsRNA is a true RNA
i inducing factor. Parrish et al. [
20] supported this discovery by using the
unc-22 gene to reveal that dsRNA functioned as a trigger to induce the RNA
i pathway. Either sense or antisense RNA strands of the target gene were not as effective as dsRNA to trigger the RNA
i mechanism. However, the double-strands did not necessarily have to be 100% homologous to the target gene. It was shown that at least 88% identity (41 bp uninterrupted identity) was required between the dsRNA trigger and target RNAs to induce gene silencing.
MECHANISMS OF RNAi
The process of RNA
i can be divided into 2 phases; initiation phase and effector phase. In the initiation phase, long dsRNA is processed into short 21-23 bp RNA by dicer, a ribonuclease III enzyme which generally leaves 2 bp- 3' overhangs. These overhangs with 5' -phosphate groups are important for the small interfering RNA (siRNA)-induced silencing complex assembly [
21,
22]. These siRNAs assemble into complexes with additional proteins and act as triggers of the RNA
i pathway.
In the effector phase, processed siRNAs along with dicer, form the RNA-induced silencing complex (RISC). Only one dicer was found in
C. elegans, while there are two dicers, dicer-1 and dicer-2, in
Drosophila melanogaster. While both dicers are involved in siRNA-RISC assembly, only dicer-1 is required for mRNA cleavage in
Drosophila [
23,
24].
There are 2 RISC precursors in
Drosophila: R1 and RLC. R1 containing Dicer-2-R2D2-siRNA, is an RNA initiator complex that is ATP-independent and a precursor to the R2 and R3 complexes. R2 activity is downstream of R1 function during siRNA-dependent RISC assembly and R3 is the RNA
i effector complex [
25]. RISC loading complex (RLC) is ATP-dependent and contains Dicer-2 and R2D2 [
26].
There are various types of proteins involved in RISC formation, and research has mainly focused on 2 key molecules; dicer that is associated with the initiation phase, and Argonaute (Ago) that is associated with the effector phase. These 2 proteins interact physically in various species including
D. melanogaster,
C. elegans and humans [
27-
29]. In addition R2D2, a
D. melanogaster dsRNA binding protein, is also implicated in RISC formation, and acts as a bridge between the 2 phases. RISC formation is not effective without R2D2, although it does not affect the enzymatic activity of dicer [
30]. R2D2 is homologous to RDE-4 in
C. elegans, which is also required for RNA
i in
C. elegans [
28].
Researchers focused on genes that connected RNA
i to the silencing of transgenes or viral activity, and concentrated on the Ago family [
31-
33]. Ago proteins play important roles in RNA
i because siRNA molecules bind to Ago proteins and serve as guides to the target mRNA [
34-
36]. Ago proteins contain 2 domains, PAZ (Piwi / Argonaute / Zwille) and Piwi (P-element induced wimpy testis) which are involved in RNA interference activity [
37,
38].
The RNA
i mechanism cannot function unless Ago proteins are intact. A mutation in
Ago1 in
Arabidopsis revealed the protein was essential for post-transcriptional silencing of transgenes [
32]. Deletion of endogenous
TbAgo1 in
Trypanosma brucei abolished RNA
i silencing in a trypanosoma cell line. This result suggested that the Ago protein is required for RNA
i in
T. brucei [
39].
The relationship between the Ago protein and RNA
i has also been studied in insect species.
Drosophila dAgo2 is involved in RISC formation. Research showed that suppression of the endogenous
dAgo2 gene reduced the ability to silence exogenous reporter gene expression in
Drosophila S2 cells. This suggested that the Ago protein is necessary for RNA
i [
40]. The function of the Ago2 protein has also been studied in multiple mosquito species. Silencing the
Anopheles gambiae Ago2 gene made mosquitoes more permissive to infection with o'nyong-nyong virus (ONNV) [
41]. The
Ago2 gene in
Aedes aegypti was also characterized. Injection of
Ae. aegypti with dsRNA targeting
Ago2 resulted in a higher titer of dengue virus type 2 (DENV-2) [
42]. These results showed that silencing the
Ago, which reduced RNA
i effects, resulted in an increase in viral titer. The authors suggested that these findings may be extrapolated to transposons because RNA
i had evolved as a defense mechanism against both viruses and transgenes [
43].
An important step after the assembly of RISC is dislodging of the sense siRNA from the RISC complex while antisense strands remain attached to the siRISC. Although this process was thought to be catalyzed by an RNA helicase [
44,
45], Matranga et al. [
46] proposed that Ago2 directly binds to siRNAs and cleaves the sense strand in an ATP-independent manner. Therefore, siRNAs enter RISC in a double-stranded form, but only the 5' -phosphorylated antisense siRNAs are chosen to act as the guide in finding the target RNA [
47]. After formation of siRISC in
Drosophila, the complex is processed to the 80S holo-RISC complex (R3), which is ready to cleave its mRNA target [
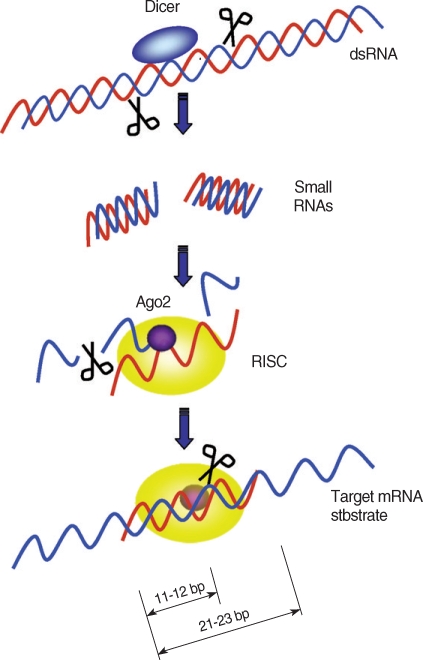
25]. Once the RISC binds to a target mRNA, a cleavage site on the mRNA-backbone is determined. The cleavage site is located at the center of the siRNA-covered region, 11-12 bp downstream of the 5' end of the nucleotides complementary to the siRNA (
Fig. 1) [
48].
The size and shape of siRNAs may be key factors in order to trigger the RNA
i mechanism efficiently. It has been known that 19-21 bp synthetic siRNAs with two-base 3' overhangs preferentially trigger RNA
i [
48]; however, there have been other studies to enhance the efficiency of RNA
i. Kim et al. [
49] showed synthetic 25-30 bp RNAs were 100 times more effective in silencing target gene expression than conventional 21 bp siRNAs. Siolas et al. [
50] showed the synthetic 29 bp short hairpin RNAs (shRNAs) with 2 bp-3' overhangs were more potent inducers than 21 bp siRNAs for RNA
i. This allows a reduction in the concentration of synthetic RNAs required to silence a target gene, which in turn allows more specific gene silencing as high doses of synthetic dsRNA may cause non-specific lethal effects.
MicroRNAs (miRNA) are another essential trigger involved in RNA
i induction. They are endogenous non-coding RNAs that are usually 22 bp long. These miRNAs enter the RISC and act as triggers of the RNA
i mechanism, similar to siRNAs. The miRNAs bind to mRNA and slow down translation. In addition, miRNAs can regulate mRNA translation, and this is believed to be an endogenous gene regulation mechanism [
51].
RNAi IN PARASITIC PROTOZOA
Trypanosomatids
RNA
i in
T. brucei has been studied extensively since the machinery was discovered in the organism [
11].
T. brucei has been used to elucidate the functions of proteins comprising the RNA
i machinery such as Ago1 and dicer [
52-
54], as well as basic RNA
i mechanisms. Researchers constructed a stem-loop that generated hairpin-loop dsRNAs targeting the α-tubulin gene under the control of the tetracycline-inducible promoter. This system inhibited α-tubulin expression in
T. brucei [
55]. Wang et al. [
56] used a vector (pZJM) in which a PCR amplified gene fragment was ligated between opposing promoters to inhibit specific gene expression in
T. brucei. Therefore, sense and antisense RNAs were simultaneously synthesized and hybridized, generating dsRNAs molecules. The synthesis of dsRNA targeting α-tubulin transcripts was induced by tetracycline.
It was confirmed that
T. congolense, which is the causative agent of nagana disease in cattle, also has RNA
i machinery by utilizing a tetracycline-regulated vector expression system [
57]. The constructed vector was transfected into
T. congolense cells, inducing the same RNA
i mechanisms as in
T. brucei. The α-tubulin synthesis was reduced when the vector was transfected into a
T. congolense cell line in the presence of tetracycline, demonstrating similar results to the previous experiment targeting α-tubulin gene in
T. brucei. Induction of the RNA
i pathway changed the morphology of transfected
T. brucei cells after α-tubulin transcripts were silenced [
11]. This suggested that RNA
i machinery exists in
T. congolense.
Conversely,
T. cruzi,
Leishmania donovani and
L. major lack RNA
i machinery although they belong to the same family as
T. brucei that contains the necessary components of the RNA
i pathway [
58-
60]. Upon searching genome databases, it was found that these species are deficient in orthologs of the Ago1 protein. It is not surprising that RNA
i has not been observed without Ago1, since this protein is required for suppression of foreign and endogenous transgenes [
32]. Another missing component is the PAZ domain, normally present in Ago. The presence of a Piwi domain is indicative of a functional RNA
i mechanism because it is a part of the Ago protein that associates with siRNA to cleave target mRNA [
61]. However, species that do not have an RNA
i mechanism lack proteins with a PAZ domain [
16], which is a subdomain of Ago and the dicer protein which binds to siRNA, contributing to firm incorporation of siRNA and miRNA into the RISC complex [
62]. The absence of these domains may explain why certain species do not have an RNA
i pathway.
Apicomplexa is characterized by its unique feature, an apical complex that projects to contact the host cell. The RNA
i pathway has been researched in
Plasmodium, but it is still controversial whether the RNA
i pathway functions in
Plasmodium or not. RNA
i-like effects were first reported in
P. falciparum [
63]. An electroporation method was employed to investigate the role of dsRNA targeting dihydroorotate dehydrogenase (DHODH), an enzyme related in the pyrimidine biosynthesis pathway. Treatment with dsRNA resulted in reduction of DHODH transcript levels, causing an inhibition of parasite growth [
63]. Another study providing evidence for the presence of RNA
i was the reduction of a cysteine protease by dsRNA in
P. falcipaum. The treated
Plasmodium showed a typical cysteine protease deficient phenotype. Moreover, authors showed that the introduced dsRNA was processed into short 25 bp fragments. Therefore, this study seemed to provide clear evidence for the presence of RNA
i machinery in
Plasmodium [
64].
However, Ullu et al. [
16] disagreed that this study provided sufficient evidence to confirm the presence of RNA
i in
Plasmodium. They argued that McRobert and McConkey [
63] did not perform a Northern analysis to confirm the data. Therefore, it was not proven that the RNA
i-induced phenotype was not a non-specific RNA
i effect. They also questioned if the siRNA found in
Plasmodium [
64] may have been broken dsRNAs of cysteine protease, resulting from contamination by the host cells [
16]. The contamination hypothesis was further supported by a subsequent paper [
65], in which short RNAs were searched to confirm that siRNA exists in
P. falciparum and eventually to provide evidence for the RNA
i pathway in
Plasmodium. However, the authors did not find any short RNAs and discovered miRNAs that turned out to be human miRNA, suggesting the results by Malhotra et al. [
64] might have been false positives [
65]. Central evidence supporting the absence of RNA
i pathway in
Plasmodium is based on the lack of canonical proteins and protein domains such as dicer, Piwi, PAZ, or RdRp that are found in the RNA
i mechanism [
16].
Database mining has hinted at the presence of classical RNA
i genes in
Toxoplasma gondii, suggesting that RNA
i could potentially exist in
Toxoplasma [
65]. A recent report suggested the existence of RNA
i pathway in
T. gondii when the introduction of dsRNA targeting uracil phosphoribosyltransferase reduced endogenous target RNA transcript levels, and this result was confirmed by Northern analysis [
66]. However, there was no evidence of siRNA inducing RNA
i in the treated
T. gondii, which makes the presence of an RNA
i pathway in
T. gondii doubtful [
16]. Some researchers argue that antisense oligonucleotides (ASO) can reduce target mRNAs in the absence of RNA
i in protozoans [
67]. In fact,
Plasmodium genes such as cytoadherence-linked gene and glucose-6-phosphate dehydrogenase gene were silenced by their antisense RNAs, suggesting that antisense RNAs can reduce their target gene transcripts levels in the absence of RNA
i machinery [
68,
69]. ASO may also explain the reduction of target transcripts in
T. gondii.
In order to search for proteins crucial to the RNA
i pathway in a specific species, database mining is quite informative. Structural proteome databases were searched and a dicer protein was identified in
Giardia intestinalis [
70]. A model was constructed in which dicer processed dsRNA into short RNAs and simulated the RNA
i pathway in
Giardia. Characterization of RNase III and Ago2, hallmarks of the RNA
i pathway, led to the prediction that RNA
i was present in
Entamoeba histolytica [
71]. Screening for the presence of siRNA in
E. histolytica is an alternative method to check for the existence of an RNA
i pathway. Ullu et al. [
72] identified a novel class of sense and antisense RNAs homologous to a retroposon family, GilT / Genie1 in
Giardia. The sense and antisense RNAs were 20 to 30 nucleotide-long and fragmented, suggesting they may inhibit the retroposon via an RNA
i pathway. This prediction should be experimentally confirmed prior to practical utilization of the RNA
i pathway in
Giardia.
It is important to first identify RNA
i genes in protozoan species because many protozoans may lack the genes responsible for the RNA
i pathway. Once the presence of the RNA
i molecules is confirmed, RNA
i can be employed to elucidate gene function or to hinder parasite development for control purposes. RNA
i can be utilized by a vector system, which would generate dsRNAs under the control of an inducible promoter or with tissue-specific expression. A vector system has been studied in
T. brucei and can be applied to different parasitic protozoan species to achieve temporal and spatial induction of RNA
i. Some researchers described the role of antisense RNA in knocking down target mRNA in
Plasmodium. Although this is not a conventional RNA
i pathway, it should be further exploited to use the antisense-knockdown system to silence specific genes [
73].
RNAi IN PARASITIC HELMINTHS
RNAi can be used to contribute to the control of parasite worms. RNAi silencing of critical genes may kill worms directly or interfere with crucial functions necessary for development. RNAi can be also used as a tool to study gene functions in helminths just as it has been employed in other species. Moreover, genes identified by RNAi can be applied as target genes for drug development or vaccine candidates, allowing utilization of RNAi for therapeutic purposes.
To date, only 10 species (8 in nematodes and 2 in trematodes) have been studied for RNA
i effects in animal parasitic helminths [
74]. This is somewhat surprising considering that the nematode,
Caenorhabditis elegans was the first organism in which dsRNAs were proven to be an RNA
i inducing factor and it has served as a model organism to characterize the RNA
i mechanism. Below are the species of parasitic helminths in which RNA
i has been studied.
Nippostrongylus brasiliensis [
75], a rat intestinal parasite, was the first nematode for which RNA
i effects were reported. This parasite is an important animal model since it shares a similar life cycle with the human hookworms,
Necator americanus and
Ancylostoma duodenale. When 1,799 bp-long dsRNAs targeting full length of acetylcholinesterases (AchEs) cDNA at the concentration of 1 mg / ml dsRNA were utilized, the target gene was suppressed nearly by 80% on the first day, but then the transcripts returned to normal levels in 4 days. By targeting AchEs with 240 bp of dsRNAs, AchEs were suppressed by more than 90% and the effects lasted for 6 days, suggesting that the short dsRNA were effective in suppression of target gene expression. This experiment provided the first evidence of a successful RNA
i effect in a parasitic helminth. Therefore, control of helminths via RNA
i seems to be feasible once a suitable target gene is identified.
RNA
i effects in
Brugia malayi suggested a more promising control method while targeting housekeeping genes (β-tubulin and RNA polymerase II large subunit) [
76]. This study showed that 300 bp long dsRNA was effective enough to result in death of the filarial worm. The authors also utilized RNA
i to target another gene, microfilaria sheath protein 1 / mf22, but this was not lethal to the worms although microfilariae release was reduced and half of the released microfilariae did not have fully elongated sheathes.
B. malayi is an important lymphatic filarial nematode and it is difficult to block their transmission by mosquitoes. Therefore, the lethal effect from RNA
i was significant as it proved to be a potential control system for
B. malayi.
Although previous studies reported a high concentration of dsRNA was needed to knock down target genes in various helminths [
75-
77]. Pfarr et al. [
78] claimed that low concentration of dsRNA was enough to specifically knock down a target gene in
Litomosoides sigmodontis, a rodent filaria. All concentrations ranging from 0.035 to 35 µM of dsRNA comparably reduced actin gene transcripts in adult worms by more than 90%. They also measured induction of hsp60 gene to figure out any stressful response on dsRNA injection. Of these concentrations, 3.5 µM of dsRNAs reduced the target with the least variation and no hsp60 induction, but high concentrations (17.5 and 35 µM) resulted in a significant increase in hsp60 transcripts, indicating stress in the filarial worm by high dsRNA dosage. This study suggests that low concentrations are enough to reduce transcript levels consistently whereas high concentrations of dsRNA may be stressful to the filarial worms. Thus, titration of appropriate concentration of dsRNAs would be required prior to RNA
i experiments.
Functional RNA
i-knockdown was reported in L3 larvae of
Onchocerca volvulus. Lustigman et al. [
79] targeted cathepsin L and cathepsin Z-like cysteine proteases that play important roles in L3 to L4 larvae for molting. Soaking the third-stage larvae (L3) in a dsRNA solution reduced the molting rate by 92% for cathepsin L and 86% for cathepsin Z-like cysteine proteases. Gene silencing of these cathepsin transcripts delayed the molting process by 1-3 days, resulting in significant reduction in the viability of the L3 larvae in
O. volvulus [
80]. Gene silencing of inorganic pyrophosphatase of the parasitic round worm,
Ascaris suum, inhibited molting from L3 to L4 by 31% [
81]. Although the inhibition rate was lower than that of
O. volvulus, discovery of functional gene silencing by RNA
i in
Ascaris worms was significant and this can be further exploited as a model system to study RNA
i in human ascariasis.
Issa et al. [
82] suggested that siRNA and electroporation are more efficient molecules and a delivery method, respectively, to induce gene silencing by RNA
i in the sheep gastrointestinal parasite,
Trichostrongylus colubriformis. The authors tested three different RNA
i delivery methods; feeding of
Escherichia coli expressing dsRNA, soaking of siRNA or dsRNA, and electroporation of siRNA or dsRNA. Ubiquitin and tropomyosin were used as target genes since their DNA sequences are well conserved and readily available. Ubiquitin transcripts were not reduced by the
E. coli feeding method, but tropomyosin was suppressed. siRNA in both electroporation and soaking resulted in a significant reduction for both target genes. This study demonstrated that electroporation is likely to result in more consistent gene silencing, and that 22 bp siRNA was more effective than long dsRNA in reducing expression of both target genes.
A variety of RNA
i conditions were tested to optimize RNA
i effects in
Haemonchus contortus, a barberpole worm. Various life stages (L1- L4 and adult), 11 genes (β-tubulin, sec-23, Ca
2+ binding protien, HSP70, vacuolar ATPase, cathepsinL, paramyosin, Cu-Zn superoxide dismutase, intermediate filament, type IV collage and GATA transcription factor) and 3 different RNA
i delivery methods (feeding, soaking and electroporation) were tested in this organism to investigate RNA
i [
83,
84]. Two β-tubulin genes that were targeted by RNA
i affected 3 life stages (L3, L4 and adult) and reduced target gene transcripts by the soaking method, however reduced motility and viability were only shown in the L3 stage [
84]. Geldhof et al. [
83] tested RNA
i effects on 11 different genes of the L1-L3 stages by 3 different delivery methods in
H. contortus. The feeding method was not effective in reducing target gene transcripts, confirming previous data in
Trichostrongylus [
82]. Only two transcripts (β-tubulin and sec-23) out of 11 genes in the L3 stage were significantly reduced by soaking in dsRNA. Interestingly, no phenotypic change was observed in the L3 larvae soaked in siRNA, and some control siRNA was even toxic to the L1 / L2 larvae. Electroporation was effective in reducing target gene transcripts in L1 larvae, as transcript levels of β-tubulin and superoxide dismutase were significantly decreased. However, larval death was observed in the L1 / L2 stage by electroporation even in the presence of control dsRNA, suggesting either electroporation is not a stable delivery method to this stage or current electroporation protocols for this stage are not optimal for analyzing RNA
i effects.
Similarly, eight genes were tested in the L1 and L3 larval stages of
Ostertagia ostertagi, a cattle parasitic nematode, by electroporation and soaking delivery methods. Significant reduction of transcripts were observed for five target genes (tropomyosin, β-tubulin, ATPase, superoxide dismutase and a polyprotein allergen) in L3 larvae, but dsRNAs of a transthyretin-like protein, a 17 kDa ES protein and ubiquitin did not reduce the target gene transcript levels. Electroporation was less effective, as only two genes (tropomyosin and β-tubulin) were successfully silenced, and these RNA
i effects were not even reproducible [
85]. This non-reproducible result indicated the RNA
i delivery method will require further optimization to achieve consistent results.
RNA
i research in trematodes has been mainly focused on
Schistosoma mansoni, blood flukes, which cause hepatosplenomegaly, hematemesis or liver failure, and may result in mortality. The first report of RNA
i in
Schistosoma targeted the cathepsin B gene [
86], an enzyme previously proposed to be responsible for degradation of host hemoglobin to digestible peptides [
87]. Shistosomes were soaked in dsRNA targeting cathepsin B and cultured for six days. The authors confirmed by RT-PCR that parasites soaked in the dsRNAs showed reduced target gene expression. Subsequent studies suggested electroporation as an alternative way to introduce dsRNA, and reduction of transcript levels of cathepsin B in schistosomula was demonstrated. Later RNA
i research proved that cathepsin B is essential for parasite growth, and not essential for hemoglobin digestion [
88].
RNA
i was used to test another gene function in the sporocyst developmental stage of schistosomes. Boyle et al. [
89] knocked down SGTP1, a facilitated diffusion glucose transporter and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) by RNA
i. The expression of both genes in the sporocyst stage was reduced when miracidia, a free living-larval stage of schistosome were soaked in its homologous dsRNA and allowed to transform to the sporocyst stage. Glucose transport activity was reduced when SGTP1 was knocked down, demonstrating the function of the gene in schistosome. Interestingly, however, when dsRNAs were introduced at the sporocyst stage, the reduction was not observed, suggesting RNA
i is not effective when introduced into this developmental stage. The effect of RNA
i lasted for 28 days, which is similar to
C. elegans [
90]. RNA
i can, therefore, be introduced at the miracidia stage and the effects will last well into the sporocyst stage.
RNA
i was utilized to reduce transcripts of scavenger receptors that are known to be important for the binding of low-density lipoprotein to the surface of schistosomula and adult
S. mansoni. Because the parasites cannot synthesize sterols and fatty acids, uptake of host-lipoprotein by scavenger receptors is critical in the synthesis of biological membranes. Miracidia were soaked in dsRNA targeting the scavenger receptors for six days and this resulted in a 60-70% reduction of the target gene transcript level. Reduction of the scavenger receptors inhibited normal parasite development, showing a more rounded morphology in sporocysts and a shorter length in larval size [
91].
RNA
i was also used to target the gynecophoral canal protein SjGCP, in
Schistosoma japonicum, a major human pathogen in the Far East [
77]. Schistosomes cannot develop without correct signaling during sexual pairing [
92], so disruption of pairing would be good target in order to control schistosomes. It is known that
S. mansoni GCP, an ortholog of SjGCP, is related to pairing and produced only by males [
93,
94]. Cheng et al. [
77] utilized RNA
i to show that the SjGCP can be silenced in a dosage-dependent manner in
S. japonicum. The target gene transcripts were reduced by 75% with 100 nM of dsRNA, but not affected at 12.5 nM. This study suggested a potential therapeutic application of RNA
i in
S. japonicum. With reduced SjGCP, the mating in
S. japonicum may be disturbed, which will cause inhibition of parasite development in the host.
While genes are successfully silenced by RNA
i in
C. elegans, gene knockdown by RNA
i has been either impossible or inconsistent in other helminths [
74]. For example, Geldhof et al. [
83] tested efficiency of RNA
i silencing in the strongylid parasitic nematode,
H. contortus, using three different dsRNA delivery methods- feeding, soaking, or electroporation. In this study, no RNA
i was observed with dsRNA feeding among four genes tested and only two genes out of eleven, β-tubulin and sec-23, showed specific gene silencing by the soaking method. Similarly, two genes out of four had knockdown of target transcripts by electroporation of respective dsRNAs. Moreover, extents of gene silencing by RNA
i also varied in the cattle parasitic helminth,
O. ostertagi and the results were often difficult to reproduce [
85]. Disagreement of results of RNA
i in parasitic helminths may be due to different delivery methods employed for introducing dsRNAs into helminths [
83]. There appears to be no consensus as to which is the most efficient delivery method to induce RNA
i gene silencing in parasitic helminths Several reports have indicated electroporation was an efficient alternative to the soaking method in delivering dsRNA in
T. colubriformis and schistosomes [
82,
88,
95]. In contrast, others suggested that electroporation was not effective in
O. ostertagi and in fact lethal to certain stages of
H. contortus [
83,
85]. Feeding dsRNA is largely inefficient in reducing transcript levels of target genes except for tropomyosin in
T. colubriformis [
82]. Soaking is another popular method for gene silencing, but this method was less efficient for the L1 / L2 stage of
H. contortus [
83] and for
S. mansoni [
95]. Apparently, all major methods commonly used for RNA
i in
C. elegans have their limitations in the delivery of dsRNA into parasitic helminths.
The disagreement in RNA
i efficiencies among parasitic helminths may result from the absence of
sid (systemic RNA interference-
deficient)-
1,
sid-2 or
rsd (RNA
i spreading
defective)-
4 in some of parasitic helminths. SID-1, SID-2, and RSD-4 proteins are involved in cellular uptake and spread of dsRNA in
C. elegans [
96]. SID-1 is a transmembrane protein that is required for RNA
i uptake into cells and spread between cells. Therefore, the absence of these proteins in certain helminths can prevent externally-provided dsRNA from entering cells, making worms refractory to RNA
i. In
sid-1 mutant
C. elegans, systemic spread of siRNA molecules was defective with any dsRNA delivery methods (feeding, soaking and microinjection) [
97]. SID-2 is also a transmembrane protein, of which expression is limited in the apical membrane of the intestinal lumen. Therefore, SID-2 is believed to be required for dsRNA uptake from the lumen to cells but not for spreading siRNA between cells. In
sid-2 deficient
C. elegans resistant to RNA
i by soaking and feeding methods, dsRNA delivered to the pseudocoelom (body cavity) by microinjection could initiate RNA
i and siRNA were autonomously spread into cells [
98]. As a result,
sid-2 deficient
C. elegans became susceptible to RNA
i, showing reduction of target transcripts. Likewise,
rsd-4 mutant
C. elegans resistant to RNA
i by feeding was reverted to be susceptible to RNA
i by supplying external dsRNA into the pseudocoelom using microinjection [
99]. Therefore, it seems there is functional overlapping between
sid-2 and
rsd-4 proteins for systemic spread of siRNA.
Many parasitic animal nematodes are deficient of
sid-1,
sid-2, or
rsd-4 orthologs, or their homologs do not share the same functions with their counterparts in
C. elegans [
96,
98,
99]. The lack of sequence or functional conservation of
sid-1,
sid-2, or
rsd-4 may account for the discrepant RNA
i results between
C. elegans and animal parasitic nematodes. To contradict this explanation, despite the fact that there is no identifiable
sid-1 ortholog in
B. malayi, a functional RNA
i mechanism was observed by a soaking method [
76]. This suggests that there may be alternative molecules or pathways substituting
sid-1 for systemic spread of siRNAs in
B. malayi. Therefore, it remains to be seen how systemic RNA
i was achieved without
sid-1 orthologs in
B. malayi.
To achieve gene-specific phenotypic knock-downs for functional studies and therapeutic applications, certain modifications or improvements for the RNA
i delivery methods may be required in animal parasitic helminths. To this end, two approaches have been recently suggested [
96]. Firstly, one may be able to trigger RNA
i by microinjection of dsRNA into the pseudocoelom of parasitic helminths if failure of RNA
i by soaking or feeding is due to the absence of
sid-2 orthologs in the gut lumen. This modification is based on the observation in
Caenorhabditis briggsae, in which unlike
C. elegans there is no
sid-2 ortholog functionally conserved to allow entry of dsRNA from the gut lumen to the body cavity. Therefore, RNA
i cannot be initiated in
C. briggsae by dsRNA using soaking and feeding methods. However, RNA
i was possible when dsRNA was supplied to the pseudocoelom via microinjection, suggesting that microinjection can bypass barriers of dsRNA uptake in the lumen, making RNA
i possible in
C. briggsae [
98,
100]. Secondly, RNA
i may be feasible in
sid-1 or
2-deficient parasitic helminths by heterologous expression of
sid-1 and / or sid-
2 of
C. elegans as shown by Winston et al. [
98]. In
C. briggsae, for example, there is no functionally conserved
sid-2 and RNA
i by soaking or feeding is thus impossible. However, transformation of
C. briggsae with
C. elegans sid-2 allowed the uptake of dsRNA from the lumen in soaking experiments, generating systemic RNA
i in
C. briggsae [
98]. As transgenesis of helminths becomes available, development of an RNA
i system through heterologous expression of
C. elegans genes for soaking or feeding may have promising applications for gene functional studies and target identification for drug discovery (
Fig. 2).
RNA
i screen is a less demanding method for drug target studies in helminths compared to forward genetic research [
15]. If, however, RNA
i machinery requires complicated dsRNA delivery process in animal parasitic nematodes,
C. elegans can be served as a model system to study gene functions. Since RNA
i machinery is well characterized in
C. elegans, this experimental system can provide a potent tool for unveiling gene functions in other nematodes via comparative genomics [
101]. The feasibility of this approach has been recently demonstrated in
B. malayi [
102]. Using sequence analyses and comparisons including gene functional studies with RNA
i knockdowns in
C. elegans, 589
B. malayi genes were identified as being critical to the survival of the filarial worm, representing potential targets for antifilarial drug discovery. Interestingly, among those 589 genes, 10 out of top 40 candidates were already previously known to be promising targets for the drug discovery because of their roles in molting, central metabolism, and structural components. This method may be realistic only for animal parasitic nematodes whose genomes are fully sequenced such as
B. malayi [
103]. Thus, genome sequencing of important helminths should be a prerequisite for this model animal based drug target studies.
RNAi IN INSECT VECTORS
RNAi in model insect species
RNA
i has been used to silence target genes or analyze gene functions in many insect species. RNA
i induced by dsRNA was shown in
Drosophila both in vivo and in vitro [
9,
13]. By abolishing
engrailed (en) transcripts with RNA
i, Marie et al. [
104] studied additional functions of the
en gene that was previously known to control topography of axonal projections in
D. melanogaster. Using larvae injected with
en dsRNA, a change in axonal branching and synaptic outputs was found, thus demonstrating
en controls synaptic choice as well as axonal projections.
After successful trials of RNA
i in
Drosophila, the RNA
i technique was applied to other insect species. Quan et al. [
12] injected dsRNA targeting the silkworm white gene (
Bmwh3). Eggs of the wild type silkworm turn dark brown after oviposition. However, mutant strains of
Bmwh3 show different patterns, with the eggs turning light brown, or remaining white. dsRNA targeting
Bmwh3 inhibited expression of
Bmwh3, resulting in white color or a mosaic pattern of egg color. The higher the concentration of dsRNA injected, the higher frequency of white and mosaic egg production. These results support the conclusion that RNA
i could be applied to lepidopteran insects.
RNA
i exists in several mosquito species, many of which are important disease vectors. When a premembrane coding region of the dengue virus type 2 genome was expressed in C6 / 36 cells derived from
Aedes albopictus in sense and antisense orientation, the titer of specific type 2 virus was decreased, suggesting that dsRNA can induce resistance to virus infection in mosquito cells [
105]. The same resistance pathway against dengue virus type 2 was shown when the premembrane coding region of the virus was expressed in
Ae. aegypti adult mosquitoes [
106]. These experiments clearly demonstrated the existence of RNA
i in
Aedes species and suggested the possibility of the use of RNA
i to control vector-borne diseases.
An. gambiae, an important vector species for malaria, also has the RNA
i pathway [
107]. Injection of dsRNAs targeting the endogenous
Defensin gene, an antimicrobacterial peptide gene, silenced the
Defensin transcripts, which increased the activity of gram-positive bacteria in
An. gambiae. This showed the possibility of RNA
i in elucidating gene functions in anopheline mosquitoes.
Analysis of gene functions using RNA
i was employed to identify antiplasmodial genes in
An. gambiae [
108]. Microarray analysis was performed in mosquitoes infected by
P. falciparum or
P. berghei, and 11 candidate immune genes (
Tep1,
AgMDL1,
FBN8,
FBN9,
FBN39,
SPCLIP1,
APOD,
IRSP1,
IRSP5,
LRRD7, and
gambicin) were identified, and the functions of these genes were assessed by RNA
i gene silencing to show their antiplasmodium activities. dsRNAs of the 11 target genes were injected into mosquitoes and gene silencing was confirmed by real-time RT-PCR. Silencing of the each immune gene increased
Plasmodium levels. In addition, RNA
i was used to study antiviral effects of a heat shock cognate protein (HSC70B) in
An. gambiae [
109]. These reports showed RNA
i can be used to analyze gene functions in
An. gambiae.
The successful use of RNA
i was extended to another important vector species. The cell line ISE6 from the
Ixodes scapularis tick, which is a vector of Lyme disease and babesiosis, was infected by Hazara virus (HAZV) [
110]. In this study, recombinant Semliki Forest virus (SFV) carrying HAZV gene in sense and antisense orientation was used. The expression of the dsRNAs of the recombinant virus efficiently inhibited HAZV replication, as monitored by western blotting and immunofluorescence assay. This suggests that RNA
i can be used to control viral diseases and other infectious diseases in
I. scapularis.
RNA
i was recently used to examine the role of a specific gene in tsetse flies [
111]. Tsetse flies,
Glossina spp., transmit African trypanosomes (
Trypanosoma brucei spp.), but the prevalence is low because the flies are refractory to trypanosome infection. Researchers compared the transcription levels of an antimicrobial peptide gene
attacin, between refractory
G. palpalis palpalis and susceptible
G. morsitans morsitans. The levels of
attacin expression were higher in the refractory species, suggesting that
attacin may be involved in the refractoriness of
G. palpalis palpalis. Subsequently, microinjection of dsRNAs targeting
attacin was carried out in order to elucidate the role of the gene. When the
attacin was silenced by dsRNAs, the infection rate significantly increased from 20% to 40%. This study showed that
attacin is a refractory gene against African trypanosomes and also demonstrated that RNA
i can be used as a powerful tool to investigate gene functions in tsetse flies.
Injection of dsRNA in insects can induce silencing effects rapidly, but the interference is transient and gene expression in later developmental stages may not be affected. Gene suppression by RNAi is also not inheritable when dsRNAs are traditionally prepared and delivered to organisms of study. However, stable and inheritable RNAi suppression was recently developed by generating transgenic insects expressing gene-specific dsRNAs. Researchers constructed a vector-based RNAi system in various organisms, which allowed in vivo RNAi that was heritable and inducible.
A heritable RNA
i system was first tested in
Drosophila, when hairpin dsRNA was prepared in vitro and injected into
Drosophila embryos carrying the
engrailed-lacZ gene. The results demonstrated significant inhibition of
engrailed-lacZ gene expression, showing that the hairpin loop dsRNAs were as efficient as linear dsRNAs that are several hundreds bp-long. In subsequent studies, the researchers constructed transgenes that produced hairpin dsRNA in vivo. Transcription of the
engrailed-lacZ gene was controlled by the Gal4 / UAS system. The Gal4 / UAS-
IRlacZ produced hairpin-loop dsRNA and inhibited
lacZ gene expression in vivo [
112]. This system produced efficient dsRNAs to silence target genes. However, the inverted repeat construct for the hairpin formation was difficult to clone and unstable, so an improved method to generate dsRNA was required.
Giordano et al. [
113] used a Gal4 / UAS system to generate both sense and antisense RNAs simultaneously in
Drosophila to overcome the drawbacks of an inverted repeat construct. Three types of plasmids were constructed using a
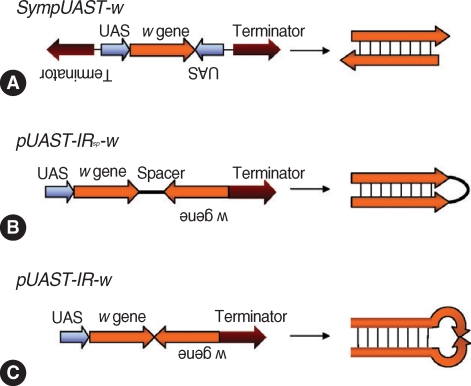
pUAST vector. First, Sym-
pUAST-w was prepared, generating dsRNA by symmetric transcription using two identical promoters (UAS) flanking the target sequence (
Drosophila white [
w] gene) in opposite directions (head-to-head) (
Fig. 3A). Then, 2 constructs,
pUAST-IRSP-w (
Fig. 3B) and
pUAST-IR-w (
Fig. 3C) were prepared to compare with Sym-
pUAST-w. Both
pUAST-IRSP-w and
pUAST-IR-w contained an inverted repeat of the
w gene, and generated hairpin dsRNAs.
pUAST-IRSP-w had a spacer between the inverted repeats while
pUAST-IR-w lacked the spacer. Three independently transformed lines of
Drosophila carrying the Sym-
pUAST-w transgene were obtained and they were mated with the
Act5C-Gal4 strain to generate hairpin dsRNA of the
w gene. The progeny had light yellow eyes, exhibiting strong silencing of the
w gene expression while the wild type had typical red eyes. The strains carrying
pUAST-IRSP-w also showed the light yellow eyes demonstrating reduction of
w expression. The progeny carrying
pUAST-IR-w showed heterogeneous expression at the phenotypic level with variegated eye color, suggesting the hairpin dsRNAs without a spacer were not efficient in silencing the target gene. This may be due to deletion in the center of inverted repeats, rendering the hairpin structure unstable. It is thus inferred that symmetrically transcribed transgenes efficiently triggered RNA
i, and the system may be effective enough to replace the inverted repeat hairpin RNA
i system.
RNA
i can be employed to test a novel method for the control of vector-borne diseases. Malaria control using germline transformation and RNA
i has been suggested [
114]. Malaria is prevalent in sub-Saharan Africa and Southeast Asia where resources are limited for disease treatments and vector control.
Historically, chemical agents were the major means to control this mosquito-borne disease. However, resistance has become prevalent among mosquitoes and parasites against insecticides and anti-malarials, respectively [
115-
119]. Therefore alternative methods to control malaria disease and transmission are in dire need. A novel method to prevent malaria transmission by anopheline mosquito vectors is attractive. Mosquito genes such as leucine rich-repeat immune gene (LRIM1), C-type lectin (CTL4) and mannose binding CTL (CTLMA2) have been identified as candidates for malaria intervention since they are critical for parasite development. Silencing these candidate genes by RNA
i resulted in impaired development of
Plasmodium ookinetes to oocysts in mosquitoes, decreasing vector competence [
114]. Therefore, this holds a potential for RNA
i to be used as a novel malaria control method.
Vector control by RNA
i can be applied to other vector species such as tsetse flies or ticks. RNA
i was used in tsetse flies to identify a refractory gene against African trypanosomes [
111], and it successfully silenced transgenes in the transfected tick cell line, ISE6, derived from
I. scapularis [
120]. These studies demonstrated the utility of RNA
i in the study of gene functions in vector-pathogen interactions. With the identification of the tick genes that are responsible for pathogen development, RNA
i can be employed to silence those genes and study their impact on pathogen development.
For gene-specific silencing, off-target effects must be considered as these non-specific effects are frequently reported in mammals [
121]. A common consequence of off-target effects in mammals is an activation of interferon dependent responses [
122,
123]. However, insects do not share the same immune pathway. Thus, typical mammalian off-target effects may not cause concerns in insect vectors. Other common examples of off-target effects of RNA
i may be over-dose lethality and toxic effects [
74]. Soaking and electroporation are the main RNA
i delivery systems in protozoans and helminths, which may cause general toxic effects due to long exposure time to dsRNA or injury by electroshock. Unlike parasitic pathogens, insect RNA
i delivery relies on mostly microinjection. Thus, off-target effects caused by injection of non-physiological amounts of dsRNAs may be preventable by careful titration of dsRNAs to be used for RNA
i experiments in insects.
CONCLUSION
RNAi techniques have the potential to revolutionize genetic manipulation and the development of therapeutic and control applications in many tropical disease pathogens and vectors. RNAi can be utilized to analyze gene functions by reducing target gene expression without altering genotypes. Thus, RNAi can help us better understand gene functions in parasites, find drug targets and vaccine candidates, or reduce the vector competence to transmit diseases. The success of this application relies on development of an optimal method for delivering a specific RNAi system into a particular organism. Soaking and electroporation have been effectively used for the introduction of dsRNAs into parasites, and a microinjection protocol has been established to deliver dsRNAs into vector insects. These provide general guidelines for performing RNAi gene silencing. However, conditions for different gene knockdowns may need modifications to suit each system or organism as varying degrees of gene silencing have been observed among different target genes and delivery methods. Conventional RNAi produces transient gene suppression effects; however, long-lasting effects may be necessary for permanent gene silencing. Germline transformation and a vector-based RNAi system would be an answer to this transient knockdown issue. Factors causing off-target effects should be identified and avoided to ensure effective gene knockdown. The cause of off-target effects may vary among different target genes, species, or RNAi delivery methods. Gene silencing by RNAi has an exciting potential to study gene functions and to suppress gene expression for disease control. To realize its full potential, the mechanisms of RNAi require further characterizations for each vector and pathogen.
References
Fig. 1Target mRNA degradation by RNAi gene silencing. Dicer initiates RNAi by cleaving dsRNAs into ~22 bp small interfering RNAs (siRNAs). During RISC assembly, Ago2 directly binds to the siRNA and cleaves sense siRNAs (blue strands) and only the anti-sense siRNAs (red strands) remain associated with the RISC complex. After assembly of RISC, the antisense strand directs RISC to target mRNAs. The RISC cut the target mRNAs at 11 to 12 bp downstream of the 5' end of the antisense strand covering the target mRNA.
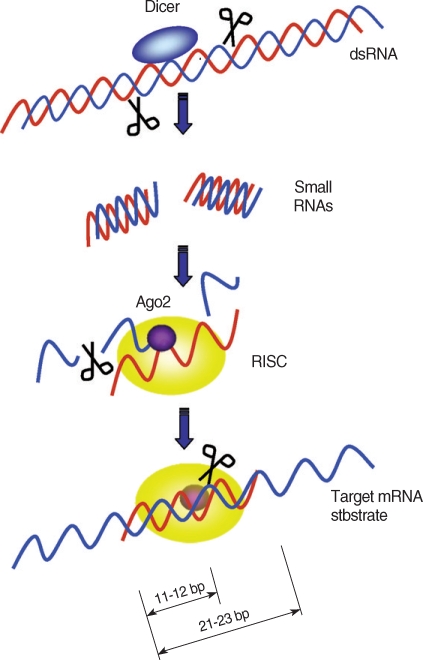
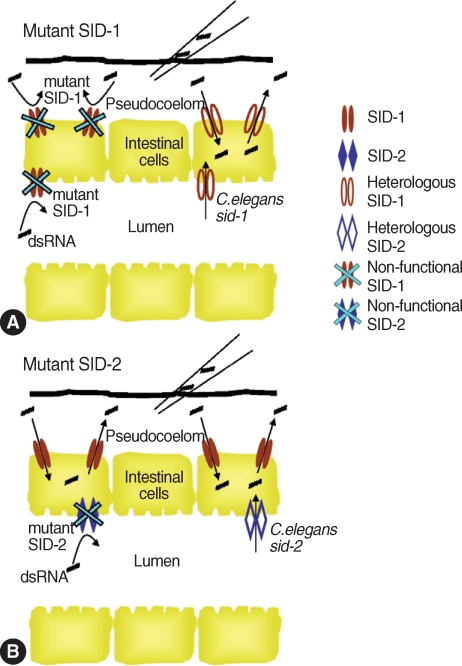
Fig. 2Functions of SID-1 and SID-2 in systemic RNA
i in
C. elegans. (A) SID-1 as a channel, allowing dsRNAs to diffuse into and between cells.
sid-1 mutant
C. elegans or
sid-1-deficient worms neither can take up dsRNAs from environment nor can spread between cells. Therefore, both microinjection and feeding methods are not effective to deliver dsRNAs. This RNA
i-deficiency can be rescued by heterologous expression of
C. elegans wild type
sid-1 [
96,
97]. (B) SID-2 also acts as a channel for dsRNAs but is only localized in the apical intestinal lumen. Thus, SID-2 is responsible for dsRNA uptake from environment (e.g. from lumen to pseudocoelom), but not for spread between cells. Feeding dsRNA is not effective for gene silencing in
sid-2 mutant
C. elegans or
sid-2-deficient worms, but microinjection can be used for dsRNA delivery because dsRNAs in the pseudocoelom can be spread systemically via
SID-1.
sid-2 mutants can be rescued for RNA
i by heterologous expression of a wild copy of
C. elegans sid-2 [
96,
98].

Fig. 3Strategies for generation of dsRNA in vivo by symmetric transcription. (A)
SympUAST-w produces dsRNAs of the
w gene by simultaneous transcription using two identical
UAS promoters flanking the target gene in opposite directions. The target gene is transcribed in both directions and the resulting sense and antisense RNAs are hybridized to form dsRNAs. (B)
pUAST-IRsp-w contains inverted repeats of the
w gene with a spacer between the repeats. This is a common approach to generate hairpin dsRNAs. (C)
pUAST-IR-w contains inverted repeats of the
w gene without a spacer. This could generate hairpin dsRNAs, but the dsRNAs were not efficient enough to silence a target gene. This may be due to deletions in the center of inverted repeats, rendering the hairpin structure unstable. it appears that symmetrically transcribed dsRNA system may be effective enough to replace the inverted repeat hairpin RNA
i system [
113].