Abstract
Clonorchis sinensis habitating in the bile duct of mammals causes clonorchiasis endemic in East Asian countries. Parkin is a RING-between-RING protein and has E3-ubiquitin ligase activity catalyzing ubiquitination and degradation of substrate proteins. A cDNA clone of C. sinensis was predicted to encode a polypeptide homologous to parkin (CsParkin) including 5 domains (Ubl, RING0, RING1, IBR, and RING2). The cysteine and histidine residues binding to Zn2+ were all conserved and participated in formation of tertiary structural RINGs. Conserved residues were also an E2-binding site in RING1 domain and a catalytic cysteine residue in the RING2 domain. Native CsParkin was determined to have an estimated molecular weight of 45.7 kDa from C. sinensis adults by immunoblotting. CsParkin revealed E3-ubiquitin ligase activity and higher expression in metacercariae than in adults. CsParkin was localized in the locomotive and male reproductive organs of C. sinensis adults, and extensively in metacercariae. Parkin has been found to participate in regulating mitochondrial function and energy metabolism in mammalian cells. From these results, it is suggested that CsParkin play roles in energy metabolism of the locomotive organs, and possibly in protein metabolism of the reproductive organs of C. sinensis.
-
Key words: Clonorchis sinensis, parkin, ubiquitin ligase, platyhelminth, mtabolism
INTRODUCTION
Clonorchis sinensis is a fish-borne trematode causing an endemic disease in East Asian countries such as Korea, China, and northern Vietnam. About 35 million people are estimated to be infected with this trematode [
1]. The adult flukes inhabit in the intrahepatic bile ducts of the fish-eating mammals. When the mammals consume infected freshwater fish, the metacercariae excyst in the duodenum of these mammals. The excysted larvae migrate through the ampulla of Vater and common bile duct and finally reach the intrahepatic biliary duct. Adult worms commonly parasitize in the intrahepatic ducts and provoke pathologic changes in the biliary tracts. This fluke provokes, in some cases, cholangiocarcinoma [
2-
4].
From the
C. sinensis transcriptome and genome, many genes were characterized to play important roles in
C. sinensis metabolism and/or pathogenesis to the hosts, but not parkin gene yet. Parkin is a causative factor of autosomal recessive juvenile Parkinsonism characterized by the loss of dopaminergic neurons in substantia nigra with symptoms such as tremor, rigidity, and bradykinesia [
5]. The 465-residue human parkin is a RING-between-RING (RBR) protein consisting of 5 domains: N-terminal ubiquitin-like domain (Ubl), 3 RING domains, and an in-between-ring domain (IBR). The RING domains have each 8 cysteine/histidine residues in the general formula C-x
2-C-x
(9-39)-C-x
(1-3)-H-x
(2-3)-C-x
2-C-x
(4-48)-C-x
2-C, which bind 2 Zn
2+ ions and form a cross-brace, which does not resemble the classical zinc fingers [
6]. The IBR domain connects the RING1 and RING2 domains [
7-
9].
Parkin has been identified in diverse cellular processes using substrates such as α-synuclein [
10], synphilin-1 [
11], Sept5 [
12], Pael-R [
13], cyclin E [
14], α/β tubulin [
15], and JTV1 [
16], suggesting indirect transcriptional and metabolic regulations. Parkin possesses ubiquitin ligase activity that catalyzes transfer of ubiquitin conjugated with E2 to specific protein substrates [
17]. This is an important post-translational modification process of proteins. The ubiquitinated proteins are processed either to proteolytic degradation or to intracellular location. Parkin regulates internalization and degradation of epidermal growth factor receptor (EGFR) through ubiquitination of Eps15 [
18]. Parkin ubiquitinates TDP-43 and facilitates its cytosolic accumulation [
19]. Dysfunction of parkin results in accumulation of its substrate which may lead to unfolded protein stress and cell death. In addition, parkin reduces oxidative stress in mitochondria and maintains homeostasis [
20,
21]. In the genome databases, the parkin genes identified are of humans, rats, mice, and
Drosophila [
22,
23].
This study was carried out to identify C. sinensis parkin (CsParkin) on a cDNA clone retrieved from the C. sinensis transcriptome and to analyze its molecular structures and biological characteristics.
MATERIALS AND METHODS
Ethics statement
BALB/c mice (female, 7-week-old) and rabbits (New Zealand White, male, 2.2-2.5 kg) were handled in an accredited Chung-Ang University animal facility (accredited unit, Korea FDA; unit no. 36). Approval for animal experiments was obtained from Institutional Animal Care and Use Committee of Chung-Ang University (permission no. CAU-2011-0013). This study was carried out strictly in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the United States National Institutes of Health. Freshwater fish Pseudorasbora parva were purchased from a city market in Shenyang, Liaoning Province, P. R. China. This activity did not require a specific permission, as it did not involve any endangered or protected species in the field.
Bioinformatics analysis, structure modeling, and phylogenetic tree construction
An EST clone encoding a polypeptide homologous with parkin of other animals was found through analysis on the
C. sinensis transcriptome database using bioinformatics tools. This EST clone (CSA08099) was retrieved from
C. sinensis transcriptome glycerol stock [
24] and was sequenced further to reach poly (A) tail. On this cDNA sequence, putative polypeptide sequence with open reading frame was predicted using ExPASy translate tool (
http://web.expasy.org/translate/). BLASTx tool was used to search CsParkin homologues in the NCBI database. CsParkin polypeptide was aligned with parkin of other animals using Clustal X and curated manually in GeneDoc [
25], then functional domains of CsParkin were identified. The secondary and tertiary structures of CsParkin were predicted on a template rat parkin [
26] using Phyre2 [
27]. A phylogenetic tree was drawn with 24 parkins retrieved from GenBank using Neighbor-Joining algorhism in Mega 5.2 software [
28]. Bootstrap value was of 1,000 replicates.
The coding region of CsParkin cDNA was amplified by PCR using specific forward (5´-CAGATACGGAATTCATGCTTGTACGGATCTCTCAC-3´) and reverse (5´-CAGATAGT CTCGAGTTAGAGCCAATGTAAGGCTTG-3´) primers containing EcoRI or XhoI restriction enzyme site. Amplification was started with denaturing DNA at 95˚C for 5 min then followed by amplification phase of 30 thermal cycles (95˚C for 1 min, 65˚C for 30 sec, 72˚C for 2 min) and a final extension at 72˚C for 10 minPCR product was purified using a spin column (QIAGEN, Hilden, Germany), digested with the restriction enzymes and subcloned into an expression plasmid vector pGEX-4T-1. In this expression construct, recombinant CsPArkin is produced as a fusion protein to a tag protein Sj26GST. Escherichia coli BL21 (DE3) pLysS (Invitrogen, Carlsbad, California, USA) was transformed with the expression plasmid DNA. The transformed E. coli were liquid-cultured overnight at 18˚C and recombinant CsParkin protein was induced by adding isopropyl β-D-thiogalactopyranoside (IPTG) at a final concentration of 0.2 mM with supplementation of 1 mM ZnCl2. E. coli was harvested by spinning and homogenized in 1×PBS by sonication at 1 sec intervals for 10 min (35 vibrations/pulse). After spinning, the supernatant was saved and loaded onto a glutathione sepharose 4B column (GE Healthcare, Uppsala, Sweden) at 4˚C for 3 hr. Recombinant CsParkin was cleaved from sj26GST tag protein by thrombin treatment overnight at 4˚C and eluted with 1× PBS. The recombinant CsParkin protein was subjected to 10% SDS-PAGE. Protein concentration was determined using a protein assay kit (Bio-Rad, Hercules, California, USA).
Production of partial CsParkin
To produce sufficient amount of protein for mouse immunization, recombinant CsParkin protein was produced in a partial form in
E. coli. B cell epitopes were predicted on the CsParkin polypeptide sequence using IEDB program (
http://tools.immuneepitope.org/tools/bcell/iedb_input). A fragment of high immunogenicity (estimated molecular mass 21 kDa) was selected and its coding cDNA region was PCR-amplified using specific forward (5’- GACTCGGAATTCTTGCAACTAAGGTTCGATG-3’) and reverse (5’-AGATCTCGAG TCCACTCTCCAAGTACTCC-3’) primers containing
EcoRI or
XhoI restriction enzyme site. Amplicon was purified and digested with the restriction enzymes and was subcloned into pET-23a (+). After transforming
E. coli BL21 (DE3) pLysS, the partial recombinant protein was induced using 0.1 mM IPTG for 4 hr at 37˚C.
E. coli cells were harvested and lyzed in 8M urea by sonication. After spinning, supernatant containing partial CsParkin protein was loaded on Ni-NTA agarose (QIAGEN) column at a denaturing condition and stood at room temperature for 3 hr. The Ni-NTA agarose column was washed twice with 8M urea buffer and recombinant protein was eluted with 8M urea buffer containing 100 mM imidazole. The partial recombinant CsParkin protein was dialyzed against 1x PBS and checked for quality by 10% SDS-PAGE.
Female, 7-week-old BALB/c, mice were purchased from an animal supply company (Samtako BioKorea Co., Osan, Korea). For primary immunization, recombinant partial CsParkin protein (50 μg per mouse) was emulsified with Freund’s complete adjuvant and injected into the mouse peritoneum. Two weeks later, mice were injected intraperitoneally with a booster dose of 50 μg recombinant protein emulsified with Freund’s incomplete adjuvant. Blood was taken from eye fundus of the immunized mice 2 weeks after the booster injection and anti-CsParkin sera were separated from the blood.
Metacercariae and adults of C. sinensis
Freshwater fish, P. parva, the second intermediate host of C. sinensis, were digested with artificial gastric juice (0.8% pepsin, 0.8% HCl). After sedimenting and washing, C. sinensis metacercariae were collected from sediments under a dissection microscope. The metacercariae were fed twice intragastrically to rabbits with 1 week interval. Six weeks after the second infection, C. sinensis adult worms were recovered from the rabbits’ bile duct and stored at -80˚C in 1× PBS until used.
Western blotting of native CsParkin
C. sinensis adults were homogenized in 2 volumes of 1× PBS containing 1% Triton X-100 and 1× Complete™ mini (Roche, Mannheim, Germany) and kept at 4˚C overnight. The suspension was centrifuged at 13,000 rpm for 60 min at 4˚C. The second supernatant was used as a crude extract of C. sinensis adults. The recombinant partial CsParkin protein and the C. sinensis adult extract were resolved by 10% SDS-PAGE and transferred onto a nitrocellulose membrane. The membrane was blocked within 5% skim milk at room temperature for 1 hr and incubated in anti-CsParkin mouse serum at dilution of 1:100 at 4˚C overnight. After washing 3 times with PBS-Tween 20, the membrane was incubated with goat anti-mouse IgG alkaline phosphatase-conjugated antibody (Sigma-Aldrich, St. Louis, Missouri, USA) at dilution of 1:5,000 at room temperature for 2 hr. Color was visualized using a substrate BCIP/NBT (Si
gma-Aldrich).
C. sinensis total RNA and quantitative real-time PCR
C. sinensis adult and metacercariae were ground into powder in liquid nitrogen using mortar and pestle. Total RNA was extracted using TRIzol reagent (Ambion, Carlsbad, California, USA) according to the manufacturer’s instruction. From the total RNA, trace of genomic DNA was removed using DNA-free kit (Ambion). Quality of the total RNA was evaluated by determining the OD260/280 using a spectrophotometer. The first-strand cDNA was synthesized using power cDNA synthesis kit with oligo-d(T) primer (INTRON Biotechnology, Gyeonggi-do, Korea) according to the manusfacturer’s protocol.
To perform quantitative real time-PCR (qRT-PCR), primers for CsParkin were designed using the Oligo ver. 6.71 software (Molecular Biology Insights, Cascade, Colorado, USA). Three genes, i.e.,
β-actin,
calcyphosine and
phosphoglycerate kinase, expressed stably between the adult and the metacercaria [
29], were employed as reference genes. The primer sequences used for qRT-PCR are in
Table 1. Reaction mix of 10 μl was made of 70 ng total cDNA of the adults or metacercariae, primer sets each of CsParkin, β-actin, calcyphosine or phosphoglycerate kinase and 1x master mix (FasStart SYBR Green I Kit, Roche). Thermal cycle was performed using LightCycler 1.5 (Roche) as follows: the reaction mix was heated to 95˚C for 15 min, followed by 45 cycles of 95˚C for 10 sec, 60˚C for 10 sec and 72˚C for 30 sec. A developmental expression of CsParkin was calculated using 2
-ΔΔCt method and presented as fold-difference [
30].
Ubiquitination assay was performed in vitro using Parkin Auto-Ubiquitination kit (BostonBiochem, Cambridge, Massachusetts, USA). The full-length recombinant CsParkin was incubated with 1 mM Mg-ATP, 50 mM HEPES buffer, pH 7.5, 50 mM NaCl, 1 mM dithiothreitol, 11 μg/ml of human E1, 9 μg/ml of human UbcH7, and 0.1 mM of human ubiquitin at 37˚C for 1 hr. For positive control reaction, human parkin was employed as a reference from the kit. After incubation, the reaction products were deployed by 10% SDS-PAGE and transferred onto a nitrocellulose membrane. The membrane was immunoblotted using mouse anti-human ubiquitin IgG (BostonBiochem) and alkaline phosphatase-conjugated goat anti-mouse IgG antibody (Sigma-Aldrich).
Immunohistochemical staining of CsParkin in C. sinensis adults and metacercariae
C. sinensis adults in the rabbit liver and metacercariae from fish were fixed in 10% paraformaldehyde, embedded in paraffin, and sectioned into ribbons. The ribbons were deparaffinized, rehydrated, and treated with anti-CsParkin mouse sera or naive mouse sera at a dilution of 1:50 for the adult flukes or a 1:100 for the metacercariae for 2 hr at room temperature. Subsequently, the ribbons were incubated with polymer-horse radish peroxidase labeled anti-mouse IgG antibody (Dako Cytomation, Glostrup, Denmark). Color was developed using 3-amino-9-ethylcarbazole as substrate and the ribbons were counterstained with hematoxylin.
RESULTS
Molecular structure and phylogeny of CsParkin
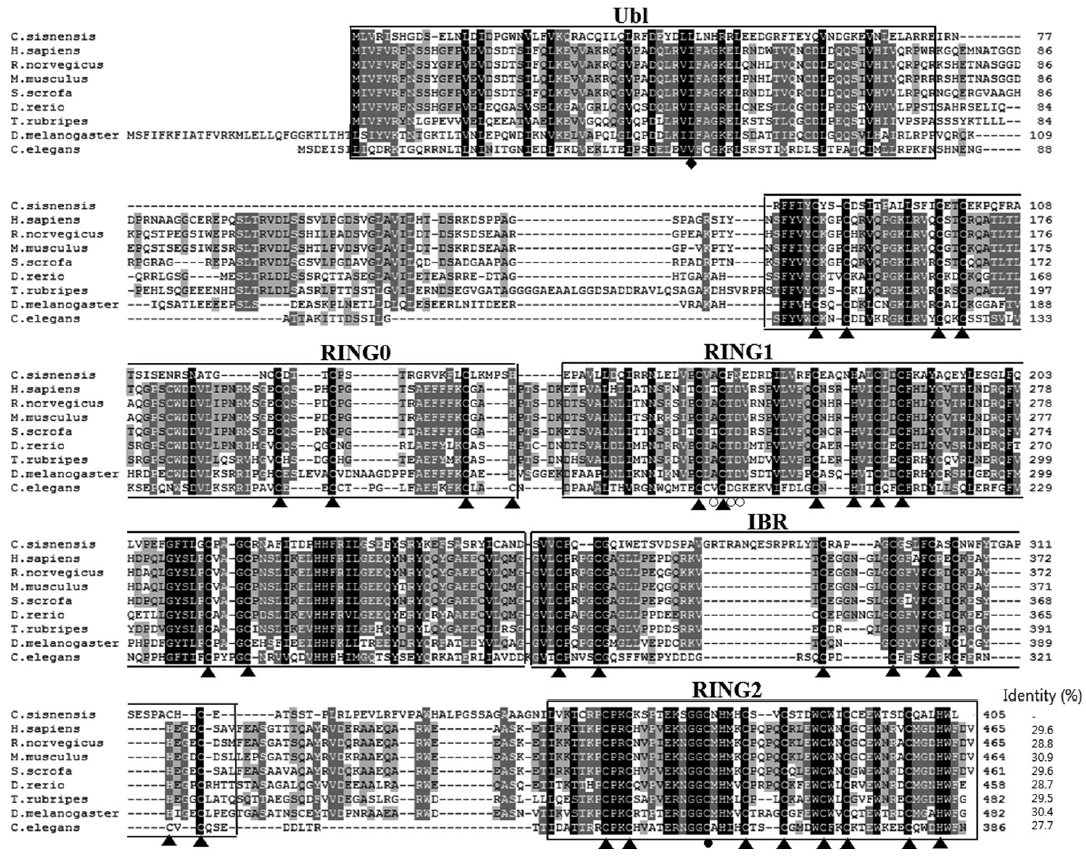
The CsParkin cDNA was 1,218 nucleotides long and encoded a putative polypeptide of 405 amino acids with an estimated molecular weight of 45.7 kDa. In the polypeptide, highly conserved were Cys and His residues which bind to Zn
2+ ion and form RING domains (
Fig. 1). Secondary structure of CsParkin revealed 5 domains in tandem: ubiquitin-like (Ubl), RING0, RING1, in-between-RING (IBR), and RING2. CsParkin lacked of a long tether between the Ubl and RING0 domains, but found in other animals. In Ubl domain, a hydrophobic RING1-binding site was conserved at Leu
43. RING1 was a canonical RING finger with classic cross-brace arrangement and had an E2-binding site consisting of Ala
164, Phe
166, and Asn
167. In all parkins, a linker between IBR and RING2, termed a repressor element of parkin (REP) [
26], displayed low similarity, but a residue Trp
339 was highly conserved. A catalytic motif consisting of Cys
375, X
376, and His
377 was well conserved and evident in CsParkin RING2 domain (
Fig. 2).
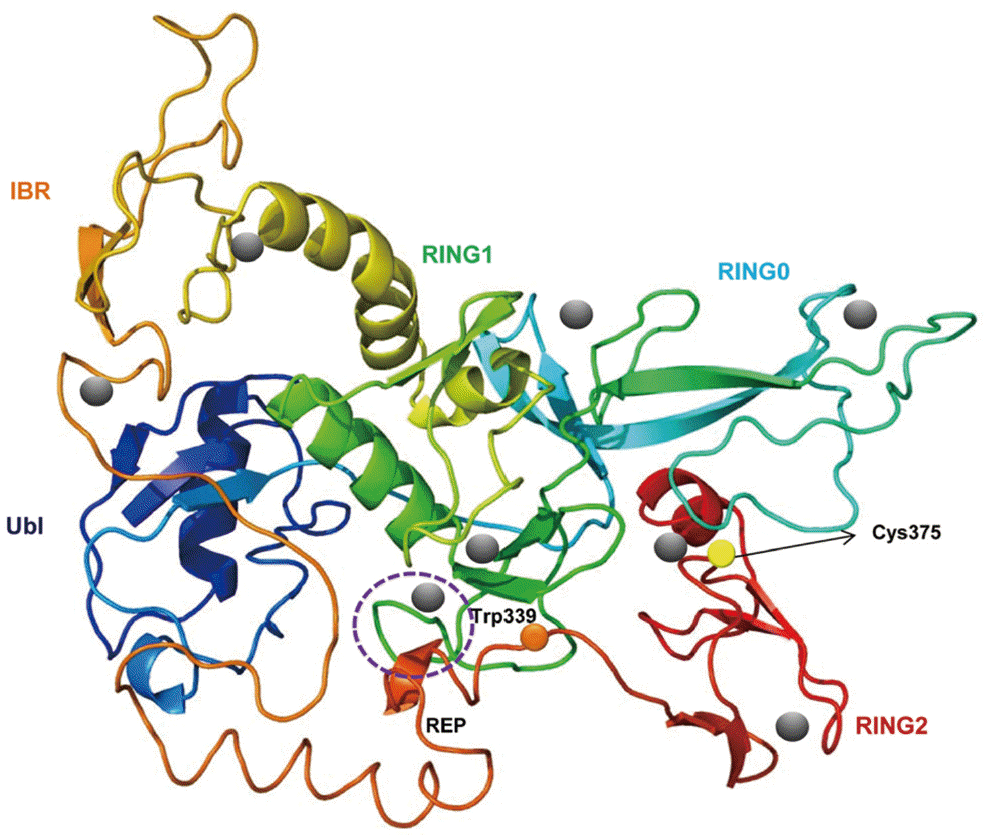
A predicted tertiary structure of CsParkin showed a very similar conformation to the crystal structure of the parkin of human and rat [
26,
31]. IBR was conformationally adjacent to RING1, while RING0 was adjacent to RING2. The E2-binding site in RING1 was occupied by REP near the Ubl binding site (
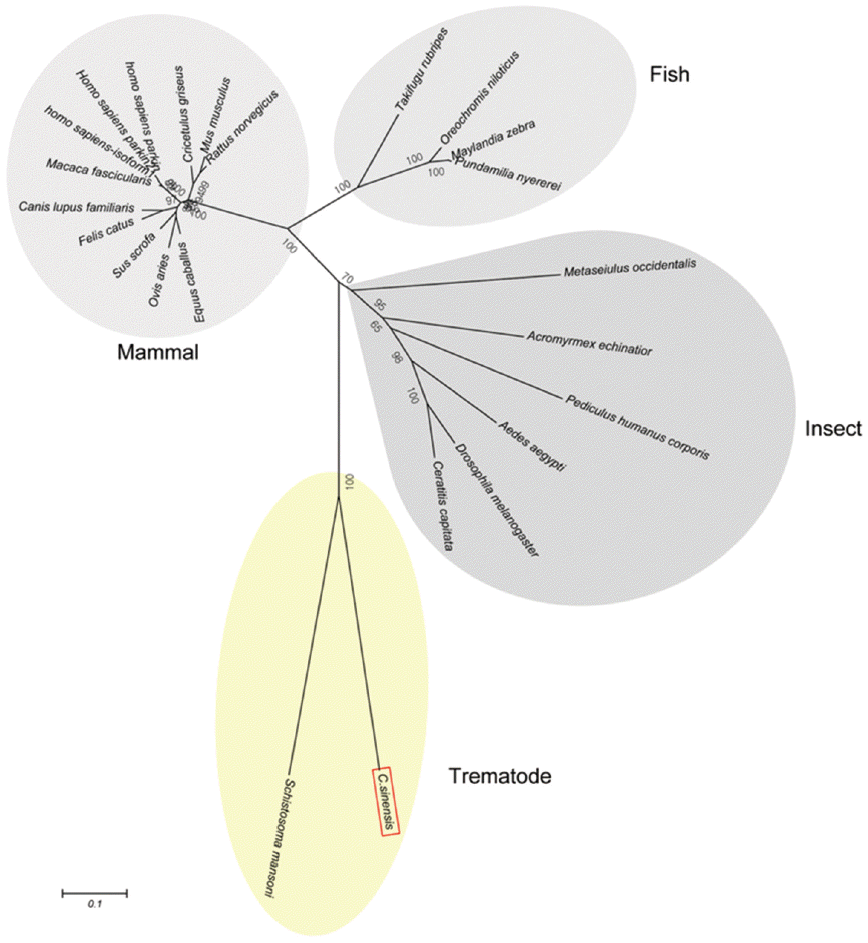
Fig. 2). A phylogenetic tree showed CsParkin grouped with a homolog of blood fluke,
Schistosoma mansoni. Parkin has not been reported from other trematodes and Platyhelminthes yet. Parkins of the mammalian animals formed a branch appearing as a major clade, followed by clades of fish and the insects (
Fig. 3).
When the recombinant full-length CsParkin fused with Sj26GST tag protein (70 kDa as whole molecule) was overexpressed in E. coli BL21 (DE3) pLysS, it remained insoluble in E. coli homogenate. Adding Zn2+ to liuid culture medium alleviate this odd and resolved a small portion of the recombinant fusion protein to soluble in E. coli. This soluble fusion protein bound efficiently to GSH-Sepharose beads. By thrombin digestion, the recombinant full-length CsParkin was on-bead cleaved from tag-protein Sj26GST and eluted as a single fraction to homogeneity. When dialyzed against PBS, part of the recombinant CsParkin remained in soluble part, but some turned to insoluble and sedimented as aggregate. This soluble CsParkin was used for its functional assay, ligase activity.
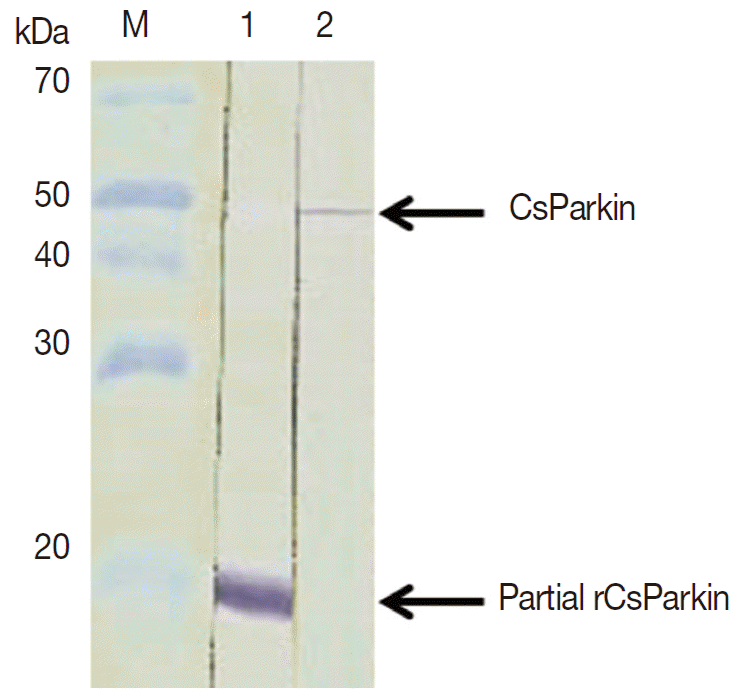
To produce a soluble form, a partial CsParkin, 21 kDa, with 6×His tag was produced in
E. coli BL21 (DE3) pLysS and purified using Ni-NTA agarose column under denaturing condition. After dialyzing against 1x PBS, the purified recombinant partial CsParkin protein remained totally in soluble fraction and near to homogeneity. This partial CsParkin was used for the production of mouse immune sera. The mouse immune sera recognized a native CsParkin showing a molecular mass close to estimated 45.7 kDa (
Fig. 4).
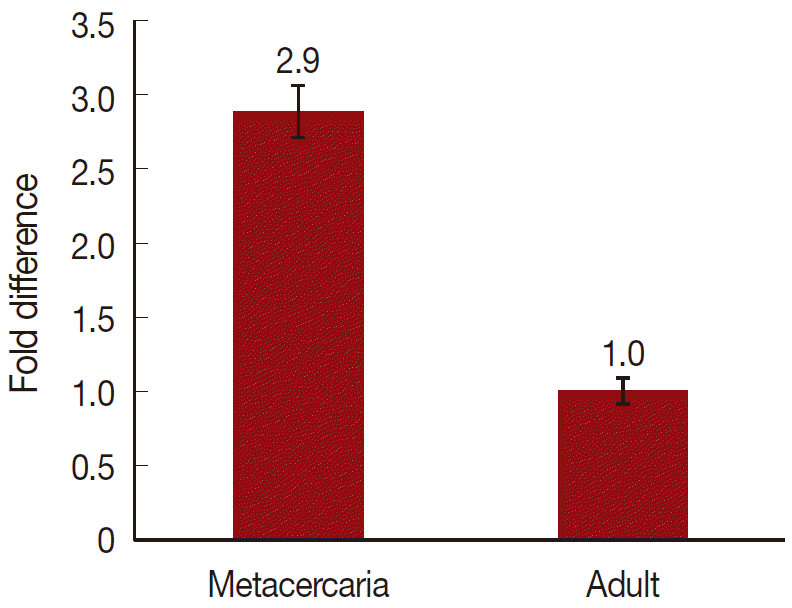
On a total cDNA as template, developmental expresion of CsParkin in the
C. sinensis adults and metacercariae was determined by qRT-PCR. CsParkin was expressed 2.9 folds higher in the metacercariae than in the adults (
Fig. 5).
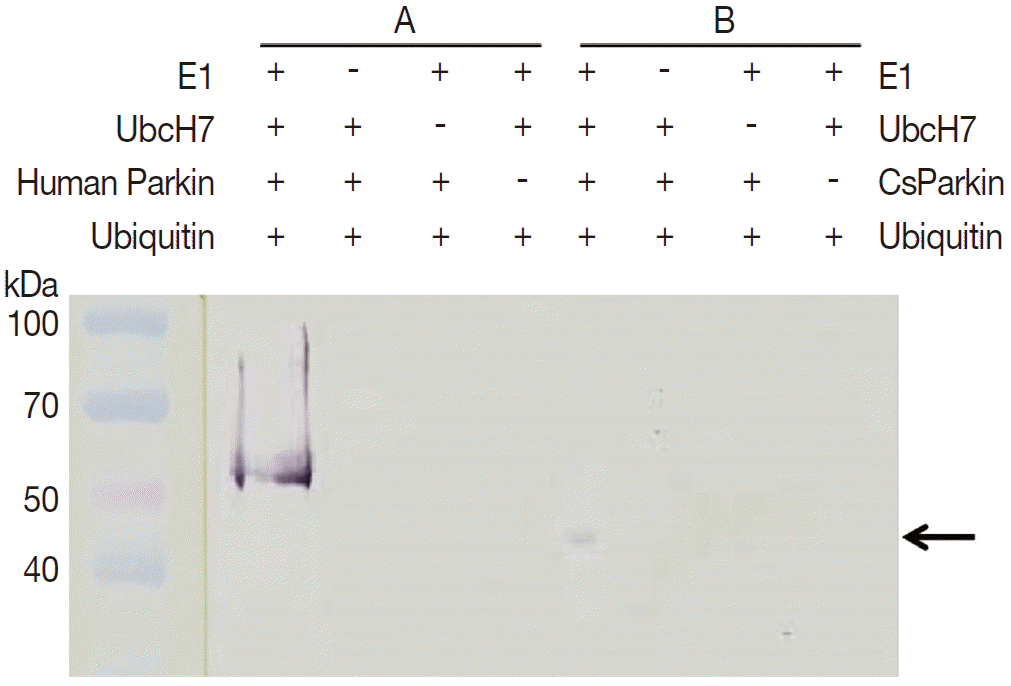
To demonstrate ubiquitin ligase activity of CsParkin, an in vitro ubiquitination assay was performed with the full-length recombinant CsParkin. In the in vitro auto-ubiquitination assay, compared to a strong band in the positive control, a small band appeared in the full-length recombinant CsParkin lane, implying that auto-ubiquitination with a heterologous human ubiquitin was taken place (
Fig. 6).
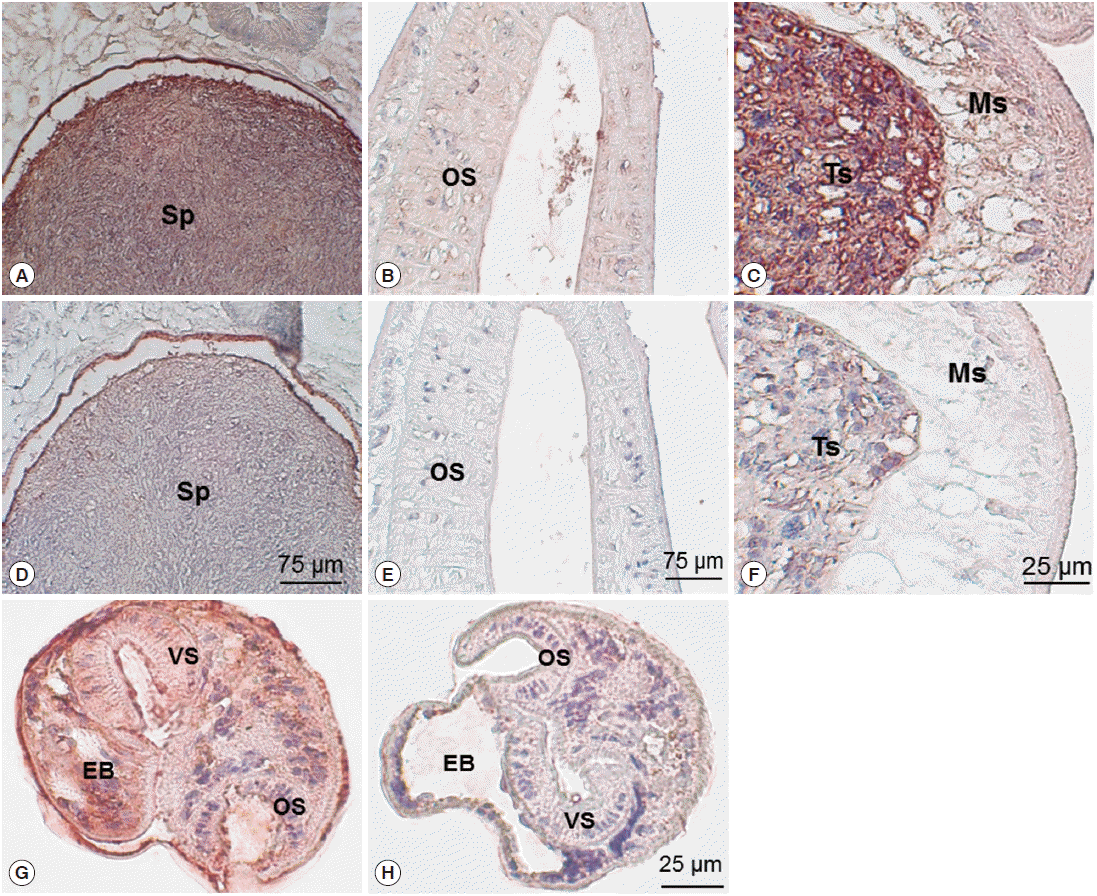
Immunohistochemical staining using mouse immune sera revealed native CsParkin in the developmental stages. In the adult worms, CsParkin was localized in the testes, sperms in the seminal receptacle and oral sucker. Weak staining was recognized in the mesenchymal tissues. In metacercariae, the CsParkin was extensively localized in the tegument, subtegument, mesenchymal tissues, and oral and ventral suckers (
Fig. 7).
DISCUSSION
Parkin has been cloned from several animals and its diverse functions have been characterized. In this study, a parkin homolog was identified from
C. sinensis and characterized for its molecular and biological functions. The putative polypeptide of CsParkin showed a high conservation of Cys and His residues, involving 3 RING domains aligned in a R0-RBR structure. The Ubl domain could bind to the S5a subunit of human 26S proteasomes or to ubiquitin-interacting motifs (UIMs) in a substrate. In both interactions, Arg
42 in the Ubl domain plays a crucial role [
18,
32]. This positively charged Arg
42 residue was replaced with negatively charged Asp
41 in CsParkin or with Glu
49 in
Caenorhabditis elegans parkin. Whether these residue replacements influence molecular function is unclear. The Ubl domain binds conformationally to the E2-binding site in the RING1 domain; this interaction was recently found in the rat parkin through crystalography [
26]. This conformation inhibits auto-ubiquitination of parkin [
33]. The RING1-binding site in Ubl domain of CsParkin included hydrophobic Leu
42 and Leu
43, very similar with other parkin homologs, suggesting an inhibitory function of CsParkin. A short tether loop between the Ubl and RING0 domains in the parkin of lower animals such as
C. sinensis and
C. elegans may allow less flexibility to tertiary structure and influence functionality.
CsParkin RING0 domain had an array of conserved (C4C3 H), as in vertebrate animals, and was predicted to coordinate with 2 Zn2+ ions. With high similarity to human parkin, the RING1 domain of CsParkin is a canonical RING finger with classic cross-brace arrangement of Cys and His residues. In the E2-conjugating enzyme binding site in the RING1 domain of CsParkin, Ala164 was highly conserved and 2 residues were substituted with Phe166 and Asn167, suggesting that CsParkin could mediate ubiquitin transfer. The IBR domain forms a pair of scissors-like and GAG knuckle-like zinc-binding sites, which bring 2 RING fingers close and facilitate protein interactions. In IBR of CsParkin, even though Cys residues were spaced longer than in those of homologues, they could flexibly bind to Zn2+ and form an IBR structure. Eventhough the REP linker between IBR and RING2 domains is dissimilar among all parkins, with one highly conserved Trp339, it plays a crucial role in repressing parkin ligase activity. Among the 3 RING domains, the RING2 domain shows the highest homology in all parkins, indicating that this region is evolutionarily conserved among all animals. As CsParkin had an E2-binding site in RING1 and catalytic residues conserved in RING2, it is suggested that CsParkin could have E3 ligase activity. The E3 ligase has dual activities of RING- and HECT-type ligases; first transfers ubiquitin from the E2 to the RING2 domain of the E3 and then to the substrate. For correct folding and structural stability of parkin proteins, Zn2+-binding to R0-RBR domains are indispensable. In producing recombinant CsParkin in E. coli, supplementation of the culture medium with Zn2+ may facilitate correct folding of the recombinant protein and retain its solubility.
The best described function of the parkin is E3 ligase activity, which interferes with ubiquitin-mediated proteolytic pathway. For CsParkin, auto-ubiquitination was detected in vitro ubiquitination assay. E3 ligase activity of CsParkin was much weaker than that of human parkin since the partner proteins added to the reaction system were heterologous, not of C. sinensis origin, which could attenuate binding of CsParkin to the substrates.
In recent years, studies have been focused on further roles of parkin in mitochondrial functionality. As an energy conversion and supplying organelle, mitochondrial homeostasis is essential to maintain cellular normality. Parkin is recruited from cytoplasm to damaged mitochondria and mediates proteasome-dependent degradation of inner and outer membrane [
34,
35]. Parkin also regulates mitochondrial spheroid formation and mitophagy via degradation of mitofusins. Mitochondrial hexokinase HK1, a pivotal enzyme involved in the cellular uptake and utilization of glucose, is a substrate of parkin, implying that parkin regulates cellular energy metabolism [
36]. Decrease in parkin expression leads to mitochondrial DNA damage and morphological change. Deficiency of parkin provokes mitochondrial dysfunction, decrease of antioxidant capacity and proportional change of proteins associated with energy metabolism [
20,
37]. In this study, CsParkin was found mainly distributed in the reproductive and locomotive organs of the
C. sinensis adults. As the parasites growing, the
C. sinensis adults produce a lot of eggs, which require large amount of energy and metabolic intermediates produced from glucose [
38,
39]. The oral sucker is a main locomotorium of
C. sinensis, reaching out and holding fast at a point then tracking the hind body. For this activity, the oral sucker, which is composed of many myocytes, consumes a great deal of energy supplied by mitochondria [
39,
40]. In the
Parkin-null
Drosophila, severe disruption of mitochondria results in apoptotic muscle cell death and deforming of sperms [
41]. In these considerations, it is suggested that CsParkin plays a role in mitochondrial energy metabolism.
CsParkin was expressed relatively higher levels and distributed more extensively in the metacercariae than in the adults of
C. sinensis. The metacercaria is a larval and resting stage with basal energy metabolism. The metacercariae has exclusively somatic organs and a genital anlage. The anlage appears as an indistinctive primodium posterior to the ventral sucker and differentiate into reproductive organs during development to adult
C. sinensis. Parkin regulates phospholipase C-γ1 and arrestin in the cell differentiation and signaling pathways [
42,
43]. Parkin can regulate EGFR through ubiquitination of Eps15, but does not bind to EFGR. Transcription levels of EGF and TGF-beta interacting protein 1 were higher in the metacercaria than in the adult
C. sinensis [
24]. It is suggested that CsParkin regulate expression of some genes, which are related to cellular proliferation and differentiation and signaling pathways in the juvenile
C. sinensis developing to the adult.
Notes
-
We declare that we have no conflict of interest related to this work.
This work was supported by the Young Investigator Research Program of Chung-Ang University (2010). We thank Mrs. Ok-Kyoung Lim for technical assistance with immunohistochemical staining.
Fig. 1.Multiple alignment of CsParkin with parkins of other animals. Conservation of amino acid residues are displayed with different background darkness; high in black and moderate in gray. An ubiquitin-like domain (Ubl) in N-terminus is boxed. RING domains are boxed and labeled with serial number. An in-between-RING domain (IBR) is boxed. The cysteine and histidine residues bind Zn2+ ions are indicated with triangles (▲). A diamond (◆) indicates RING1-binding residue in the Ubl domain. An open circle (○) denotes E2-binding site in RING1 domain. A closed circle (●) indicates a catalytic residue in RING2 domain. Aligned are parkin of Homo sapiens (BAA25751.1), Rattus norvegicus (AAG37013.1), Mus musculus (AAG13890.1), Sus scrofa (NP_001038068.1), Danio rerio (NP_001017635.1), Takifugu rubripes (AAS79348.1), Drosophila melanogaster (AAM43930.1), and Caenorhabditis elegans (CAB04599.2). Identity of each parkin protein to CsParkin is given at the end of the sequence.
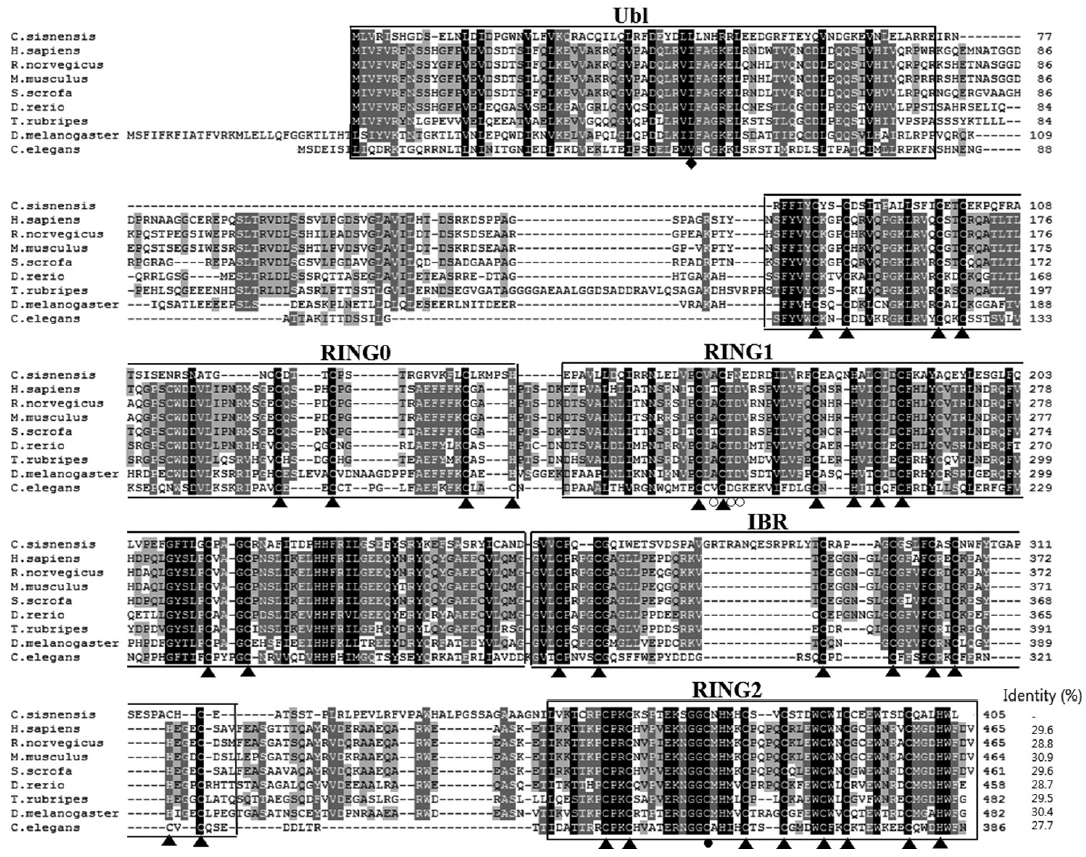
Fig. 2.A putative tertiary structure of CsParkin. The tertiary structure of CsParkin was predicted with Phyre2 using rat parkin as a template. Conformations of Ubl, RING0, RING1, IBR, and RING2 domain are displayed in different colors. A grey sphere denotes the bound Zn2+ ion. An E2-binding region in RING1 domain is marked by a broken purple circle. REP fragment containing a key residue Trp339 is identified between IBR and RING2. The catalytic residue Cys375 is depicted with a yellow sphere.

Fig. 3.A phylogenetic tree of CsParkin with homolog of other animals. Number at each node denotes bootstrap value (in percentage) generated using by Neighbor-Joining method (1,000 replicates). Parkin is labeled each with its species name. A phylogenetic tree was drawn with 24 parkin homologs of vertebrate and invertebrate animals retrieved from GenBank using MEGA5.0.
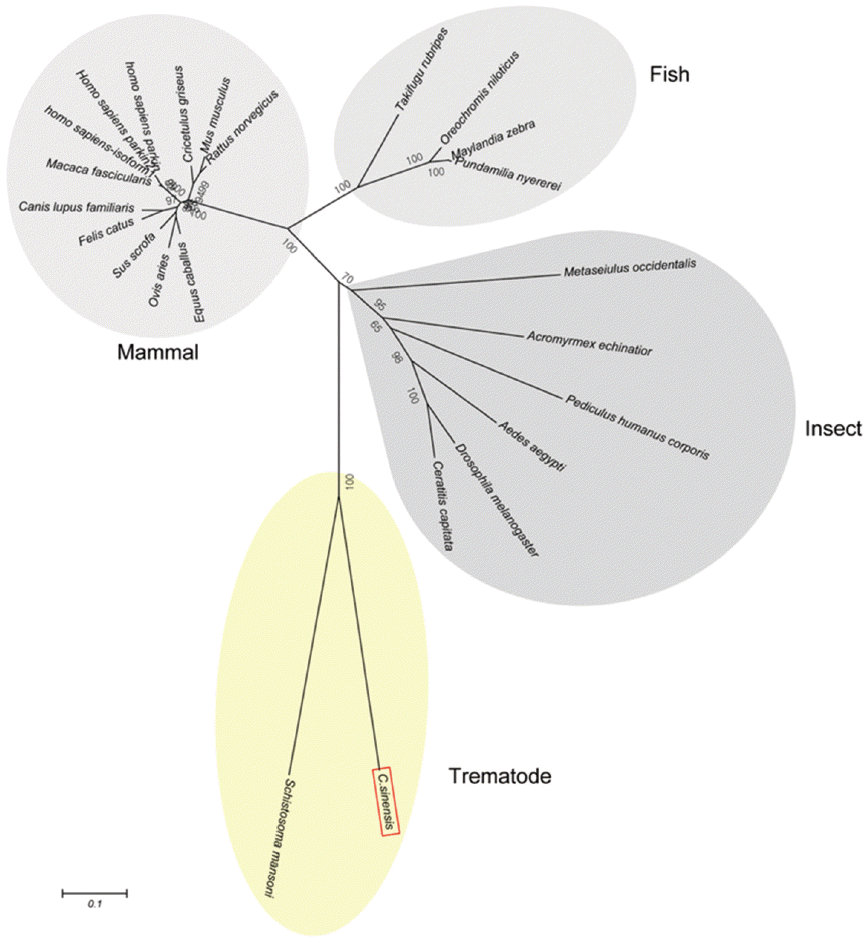
Fig. 4.Immunoblotting of partial recombinant and native CsParkins. The partial recombinant CsParkin protein and the C. sinensis adult extract were resolved by 10% SDS-PAGE and transferred onto a nitrocellulose membrane. Blots were probed with a mouse anti-partial recombinant CsParkin immune serum. Lane 1, partial recombinant CsParkin; Lane 2, native CsParkin from soluble extract of adult C. sinensis. M, molecular mass.

Fig. 5.Relative transcriptional level of CsParkin in the developmental stages of C. sinensis. Quantitative RT-PCR was performed on total cDNA of the C. sinensis adults and metacercariae. cDNAs of β-actin, calcyphosine and phosphoglycerate kinase were employed as references to calculate relative transcription of the target gene using 2-ΔΔCt equation.
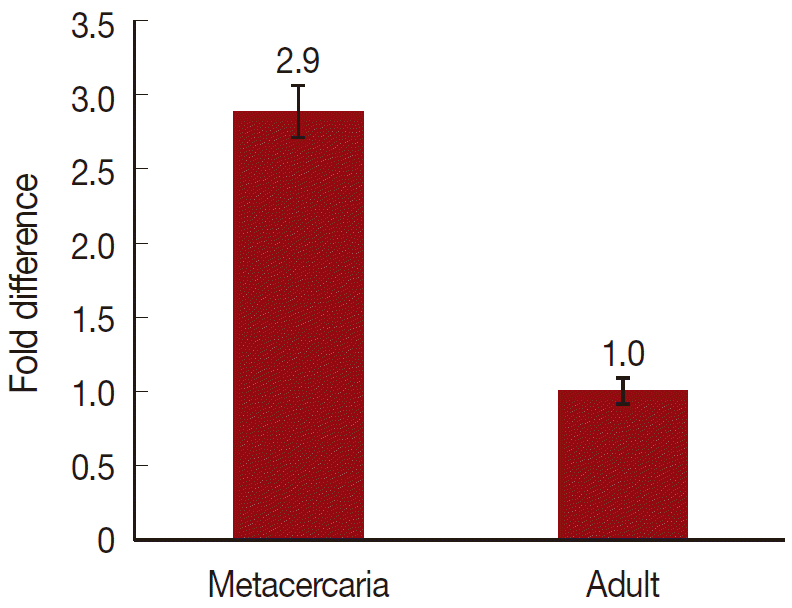
Fig. 6.In vitro ubiquitination assay on recombinant CsParkin. Full-length recombinant CsParkin was assayed in vitro for ubiquitination activity. Human parkin was employed as a positive control. Panel A, a control reaction set was run with human parkin. Panel B, the reaction set was run with recombinant full-length CsParkin. After incubation, reactions were deployed by 10% SDS-PAGE, transferred onto a nitrocellulose membrane, and probed with mouse anti-human ubiquitin IgG. An arrow indicates an ubiquitinated CsParkin.
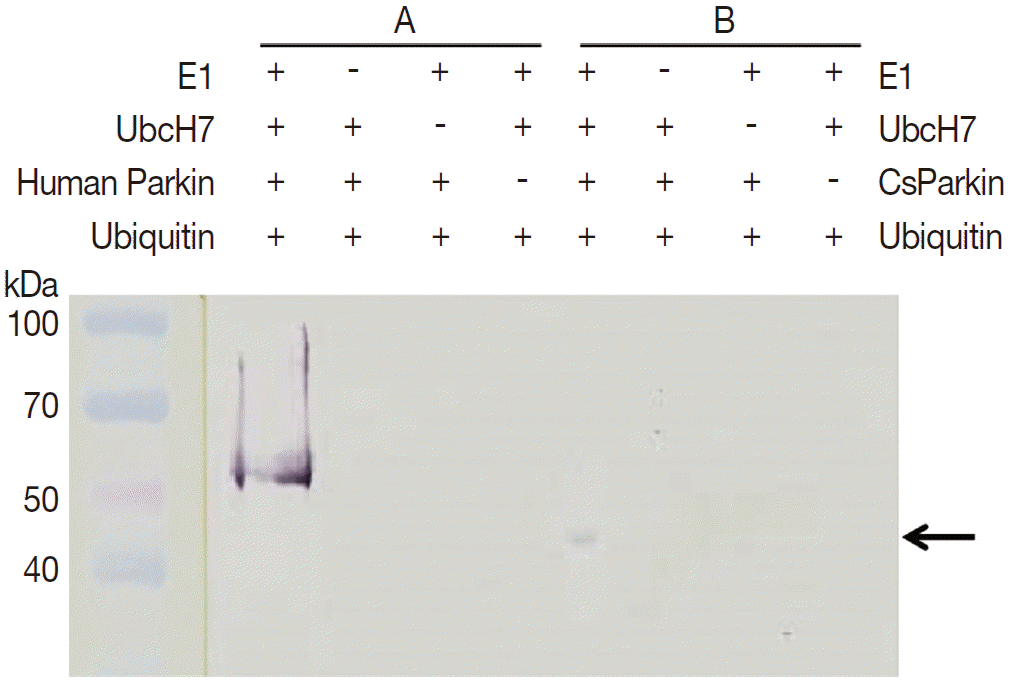
Fig. 7.Localization of CsParkin in the C. sinensis adults and metacercariae. Panels A and D, sperms (Sp) in seminal receptacle. Panels B and E, oral sucker (OS); Panels C and F, testis (Ts) and mesenchymal tissue (Ms); Panels G and H, metacercariae. Panels A, B, C, and G were probed with mouse anti-CsParkin immune sera, and Panels D, E, F, and H with normal mouse sera. For technical information, refer to the Materials and Methods.
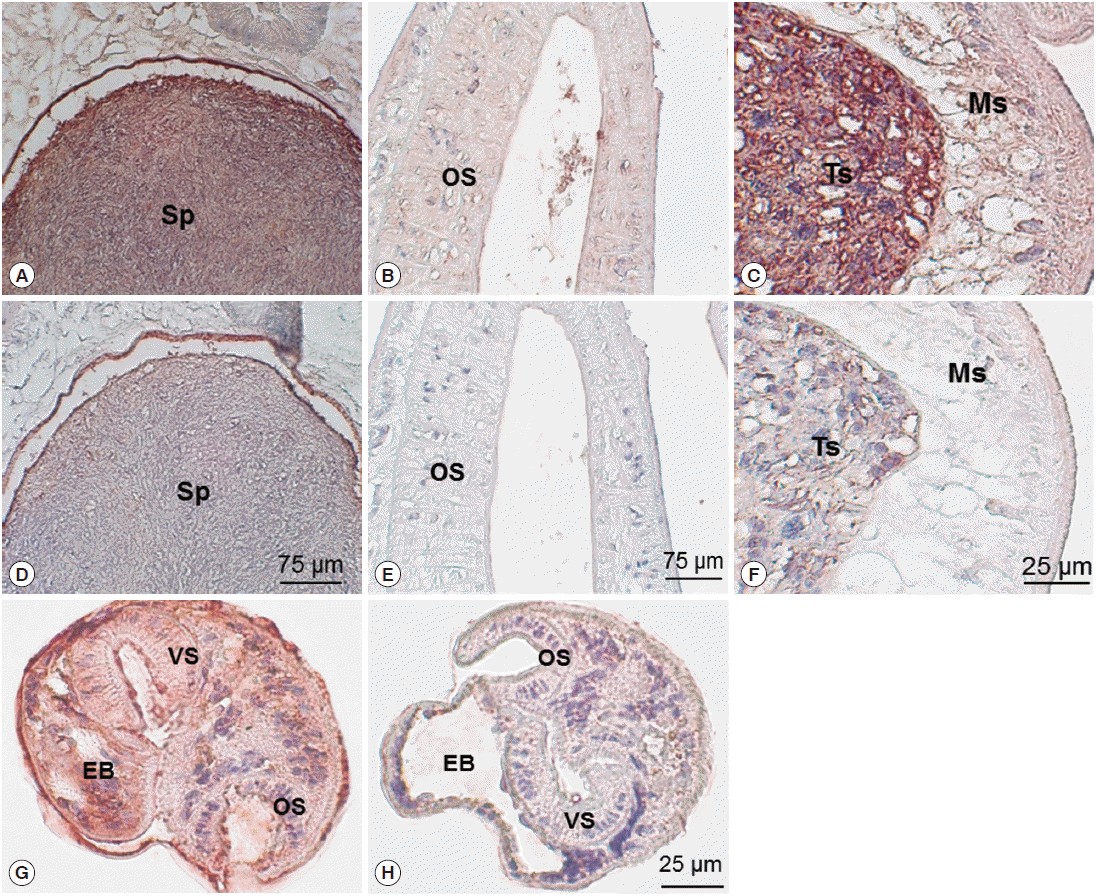
Table 1.Primer sequences of target and reference cDNAs used in quantitative real-time PCR
Table 1.
|
Target |
Forward primer |
Reverse primer |
|
CsParkin |
CACTTCCTCAACGCCATTA |
TATCCAGCACCAATCAGTAGA |
|
β-Actin |
CGCTACGATCTTGATCTTCAT |
AGTTTCCTTGGTATGGAGTCT |
|
Calcyphosine |
AATCCGAAATACCAGAACAA |
TGAACGCTTGACGAATCAT |
|
Phosphoglycerate kinase |
GCGGGTGCTTATGCGAGTTGA |
CACCGGGTTGAGGGAATATCT |
References
- 1. Lun ZR, Gasser RB, Lai DH, Li AX, Zhu XQ, Yu XB, Fang YY. Clonorchiasis: a key food-borne zoonosis in China. Lancet Infect Dis 2005;5:31-41.
- 2. Kim TI, Yoo WG, Kwak BK, Seok JW, Hong SJ. Tracing of the bile-chemotactic migration of juvenile Clonorchis sinensis in rabbits by PET-CT. PLoS Negl Trop Dis 2011;5:e1414.
- 3. Shin HR, Oh JK, Masuyer E, Curado MP, Bouvard V, Fang YY, Wiangnon S, Sripa B, Hong ST. Epidemiology of cholangiocarcinoma: an update focusing on risk factors. Cancer Sci 2010;101:579-585.
- 4. Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, Cogliano V; WHO International Agency for Research on Cancer Monograph Working Group. A review of human carcinogens-Part B: biological agents. Lancet Oncol 2009;10:321-322.
- 5. Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile Parkinsonism. Nature 1998;392:605-608.
- 6. Capili AD, Edghill EL, Wu K, Borden KL. Structure of the C-terminal RING finger from a RING-IBR-RING/TRIAD motif reveals a novel zinc-binding domain distinct from a RING. J Mol Biol 2004;340:1117-1129.
- 7. Morett E, Bork P. A novel transactivation domain in parkin. Trends Biochem Sci 1999;24:229-231.
- 8. Van der Reijden BA, Erpelinck-Verschueren CA, Löwenberg B, Jansen JH. TRIADs: a new class of proteins with a novel cysteine-rich signature. Protein Sci 1999;8:1557-1561.
- 9. Hristova VA, Beasley SA, Rylett RJ, Shaw GS. Identification of a novel Zn2+-binding domain in the autosomal recessive juvenile Parkinson-related E3 ligase parkin. J Biol Chem 2009;284:14978-14986.
- 10. Shimura H, Schlossmacher MG, Hattori N, Frosch MP, Trockenbacher A, Schneider R, Mizuno Y, Kosik KS, Selkoe DJ. Ubiquitination of a new form of alpha-synuclein by parkin from human brain: implications for Parkinson's disease. Science 2001;293:263-269.
- 11. Chung KK, Zhang Y, Lim KL, Tanaka Y, Huang H, Gao J, Ross CA, Dawson VL, Dawson TM. Parkin ubiquitinates the alpha-synuclein-interacting protein, synphilin-1: implications for Lewy-body formation in Parkinson disease. Nat Med 2001;7:1144-1150.
- 12. Choi P, Snyder H, Petrucelli L, Theisler C, Chong M, Zhang Y, Lim K, Chung KK, Kehoe K, D'Adamio L, Lee JM, Cochran E, Bowser R, Dawson TM, Wolozin B. SEPT5_v2 is a parkin-binding protein. Brain Res Mol Brain Res 2003;117:179-189.
- 13. Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, Takahashi R. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of parkin. Cell 2001;105:891-902.
- 14. Staropoli JF, McDermott C, Martinat C, Schulman B, Demireva E, Abeliovich A. Parkin is a component of an SCF-like ubiquitine ligase complex and protects postmitotic neurons from kainite excitotoxicity. Neuron 2003;37:735-749.
- 15. Ren Y, Zhao J, Feng J. Parkin binds to α/β tublin and increases their ubiquitination and degradation. J Neurosci 2003;23:3316-3324.
- 16. Corti O, Hampe C, Koutnikova H, Darios F, Jacquier S, Prigent A, Robinson JC, Pradier L, Ruberg M, Mirande M, Hirsch E, Rooney T, Fournier A, Brice A. The p38 subunit of the aminoacyl-tRNA synthetase complex is a parkin substrate: linking protein biosynthesis and neurodegeneration. Hum Mol Genet 2003;12:1427-1437.
- 17. Imai Y, Soda M, Takahashi R. Parkin supress unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J Biol Chem 2000;275:35661-35664.
- 18. Fallon L, Bélanger CM, Corera AT, Kontogiannea M, Regan-Klapisz E, Moreau F, Voortman J, Haber M, Rouleau G, Thorarinsdottir T, Brice A, van Bergen En Henegouwen PM, Fon EA. A regulated interaction with the UIM protein Eps 15 implicates parkin in EGF receptor trafficking and PI(3)K-Akt signaling. Nat Cell Biol 2006;8:834-842.
- 19. Hebron ML, Lonskaya I, Sharpe K, Weerasinghe PP, Algarzae NK, Shekoyan AR, Moussa CE. Parkin ubiquitinates Tar-DNA binding protein-43 (TDP-43) and promotes its cytosolic accumulation via interaction with histone deacetylase 6 (HDAC6). J Biol Chem 2013;288:4103-4115.
- 20. Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, Wacker M, Klose J, Shen J. Mitochondrial dysfunction and oxidative damage in parkin deficient mice. J Biol Chem 2004;279:18614-18622.
- 21. Ding WX, Guo F, Ni HM, Bockus A, Manley S, Stolz DB, Eskelinen EL, Jaeschke H, Yin XM. Parkin and mitofusins reciprocally regulate mitophagy and mitochondrial spheroid formation. J Biol Chem 2012;287:42379-42388.
- 22. Kitada T, Asakawa S, Minoshima S, Mizuno Y, Shimizu N. Molecular cloning, gene expression, and identification of a splicing variant of the mouse parkin gene. Mamm Genome 2000;11:417-421.
- 23. Bae YJ, Park KS, Kang SJ. Genomic organization and expression of parkin in Drosophila melanogaster. Exp Mol Med 2003;35:393-402.
- 24. Yoo WG, Kim DW, Ju JW, Cho PY, Kim TI, Cho SH, Choi SH, Park HS, Kim TS, Hong SJ. Developmental transcriptomic features of the carcinogenic liver fluke, Clonorchis sinensis. PLoS Negl Trop Dis 2011;5:e1208.
- 25. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics 2007;23:2947-2948.
- 26. Trempe JF, Sauvé V, Grenier K, Seirafi M, Tang MY, Ménade M, Al-Abdul-Wahid S, Krett J, Wong K, Kozlov G, Nagar B, Fon EA, Gehring K. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science 2013;340:1451-1455.
- 27. Nema V, Pal SK. Exploration of freely available web-interfaces for comparative homology modelling of microbial proteins. Bioinformation 2013;9:796-801.
- 28. Hall BG. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 2013;30:1229-1235.
- 29. Yoo WG, Kim TI, Li S, Kwon OS, Cho PY, Kim TS, Kim K, Hong SJ. Reference genes for quantitative analysis on Clonorchis sinensis gene expression by real-time PCR. Parasitol Res 2009;104:321-328.
- 30. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 2001;25:402-408.
- 31. Riley BE, Lougheed JC, Callaway K, Velasquez M, Brecht E, Nguyen L, Shaler T, Walker D, Yang Y, Regnstrom K, Diep L, Zhang Z, Chiou S, Bova M, Artis DR, Yao N, Baker J, Yednock T, Johnston JA. Structure and function of parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun 2013;4:1982.
- 32. Sakata E, Yamaguchi Y, Kurimoto E, Kikuchi J, Yokoyama S, Yamada S, Kawahara H, Yokosawa H, Hattori N, Mizuno Y, Tanaka K, Kato K. Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain. EMBO Rep 2003;4:301-306.
- 33. Chaugule VK, Burchell L, Barber KR, Sidhu A, Leslie SJ, Shaw GS, Walden H. Autoregulation of parkin activity through its ubiquitin-like domain. EMBO J 2011;30:2853-2867.
- 34. Zheng X, Hunter T. Parkin mitochondrial translocation is achieved through a novel catalytic activity coupled mechanism. Cell Res 2013;23:886-897.
- 35. Yoshii SR, Kishi C, Ishihara N, Mizushima N. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J Biol Chem 2011;286:19630-19640.
- 36. Okatsu K, Iemura S, Koyano F, Go E, Kimura M, Natsume T, Tanaka K, Matsuda N. Mitochondrial hexokinase HKI is a novel substrate of the parkin ubiquitin ligase. Biochem Biophys Res Commun 2012;428:197-202.
- 37. Periquet M, Corti O, Jacquier S, Brice A. Proteomic analysis of parkin knockout mice: alterations in energy metabolism, protein handling and synaptic function. J Neurochem 2005;95:1259-1276.
- 38. Jeong KH, Rim HJ. A study on the fine structure of Clonorchus sinensis, a liver fluke V. The mature spermatozoa. Korean J Parasitol 1984;22:30-36.
- 39. Xiao JY, Lee JY, Tokuhiro S, Nagataki M, Jarilla BR, Nomura H, Kim TI, Hong SJ, Agatsuma T. Molecular cloning and characterization of taurocyamine kinase from Clonorchis sinensis: a candidate chemotherapeutic target. PLoS Negl Trop Dis 2013;7:e2548.
- 40. Hong SJ, Seong KY, Sohn WM, Song KY. Molecular cloning and immunological characterization of phosphoglycerate kinase from Clonorchis sinensis. Mol Biochem Parasitol 2000;108:207-216.
- 41. Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci USA 2003;100:4078-4083.
- 42. Dehvari N, Sandebring A, Flores-Morales A, Mateos L, Chuan YC, Goldberg MS, Cookson MR, Cowburn RF, Cedazo-Mínguez A. Parkin-mediated ubiquitination regulates phospholipase C-γ1. J Cell Mol Med 2009;13:3061-3068.
- 43. Ahmed MR, Zhan X, Song X, Kook S, Gurevich VV, Gurevich EV. Ubiquitin ligase parkin promotes Mdm2-arrestin interaction but inhibits arrestin ubiquitination. Biochemistry 2011;50:3749-3763.