Abstract
Most of the diphyllobothriid tapeworms isolated from human samples in the Republic of Korea (= Korea) have been identified as Diphyllobothrium nihonkaiense by genetic analysis. This paper reports confirmation of D. nihonkaiense infections in 4 additional human samples obtained between 1995 and 2014, which were analyzed at the Department of Parasitology, Hallym University College of Medicine, Korea. Analysis of the mitochondrial cytochrome c oxidase 1 (cox1) gene revealed a 98.5-99.5% similarity with a reference D. nihonkaiense sequence in GenBank. The present report adds 4 cases of D. nihonkaiense infections to the literature, indicating that the dominant diphyllobothriid tapeworm species in Korea is D. nihonkaiense but not D. latum.
-
Key words: Diphyllobothrium nihonkaiense, Diphyllobothrium latum, cox1 gene
INTRODUCTION
Diphyllobothrium nihonkaiense is a parasite prevalent in Korea and Japan, but rare in China, Russia, North America, Oceania, and Europe [
1-
7].
Diphyllobothrium latum and
D. nihonkaiense are morphologically similar and difficult to distinguish.
D. nihonkaiense has been inaccurately diagnosed as
D. latum; however, this erroneous identification can be revised using DNA sequencing. Although the treatment is the same for both species [
8], the disease severity and treatment results may vary only with limited evidence. Therefore, accumulation of data is necessary.
Many case reports have indicated salmon intake as the source of
D. latum infection in Korea [
9]; however, salmon intake is now more indicative of
D. nihonkaiense infection. Other sea fish species from the Pacific Ocean [
10] may also act as intermediate hosts for
D. nihonkaiense. DNA analysis of suspected
D. latum samples collected and examined in Korea indicated the presence of
D. nihonkaiense [
11]. We performed DNA analysis of 4
Diphyllobothrium samples from patients referred to the Parasitology Department of Hallym University, Korea between 1995 and 2014 and identified them as
D. nihonkaiense.
CASE DESCRIPTION
Proglottids of Diphyllobothrium were obtained from 4 patients referred to the Parasitology Department of Hallym University, Chuncheon, Korea from November 1995 to February 2014 and stored in 100% ethanol. Only 1 patient in 2014 could be reached for medical history collection. The patient resided in Inje-gun, Gangwon-do, and frequently visited Sokcho-si, Gangwon-do to consume raw fish (=sashimi). The patient consumed raised halibut, raised rockfish, blowfish, flounder, filefish, raised mullet, sea squirt, whelk, and squid, but had no salmon intake history. The patient’s wife also consumed sashimi but not mullet because of personal preference.
In November 2012, the patient experienced repeated bouts of loose stools. The patient observed a 2-m long parasite while defecating and tried to remove it, but it ruptured. The same issue occurred 1 month later. It is notable that normal stools transformed into loose stools at the point of release of the parasite. The patient received treatment with albendazole (400 mg), after which the same parasite (a part) was excreted. He was treated with albendazole (400 mg) for 3 more days but the medication had no effects. The patient continued medication for 4 more days, with repeated excretion of the parasite. The parasite excretion lasted 1 month when only 1 parasite was observed, and the excretion started to decrease thereafter. However, in February 2014, 2-3 segments of parasites were observed during defecation. Each parasite segment was 3-4 m long, and ruptured when the patient attempted to manually remove them.
The patient visited an internist in Inje-gun with the expelled parasite. The doctor referred the patient to the Parasitology Department of Hallym University for identification of the parasite, and prescribed praziquantel (10 mg/kg in a single dose). The day after initiation of the therapy, a single dead parasite was observed in the stool. Live parasites expelled previously moved actively and were white to mild yellowish in color, whereas the dead parasite was darker yellowish in color. The proglottid but not the scolex was observed in the parasite. The patient was monitored for 3 months, with no symptoms of infection recurrence, including loose stool.
The medical histories of the other 3 patients were unavailable. Of the 4 Diphyllobothrium samples collected from patients, 3 were stored at the Parasitology Department, Hallym University College of Medicine in Chuncheon, Gangwon-do, and 1 at the Parasitology Department at Inje-gun, Gangwon-do. The 4 samples were sent to the Department of Parasitology, Chungbuk National University School of Medicine for DNA analysis.
A single proglottid was cut into small pieces, and total genomic DNA was extracted using a DNeasy tissue kit (Qiagen, Valencia, California, USA) according to manufacturer’s instructions. The partial mitochondrial cox1 gene was amplified by PCR. PCR was performed in a reaction mixture containing 50 μl with 0.01 μg/μl of genomic DNA, 10× PCR buffer (20 mM Mg+), 10 mM dNTP mixture, 10 pmoles of each primer, and 2.5 U/μl Taq DNA polymerase (High Fidelity PCR system, Roche, Mannheim, Germany) using a GeneAmp PCR System 9700 (Applied Biosystems, Langen, Germany).
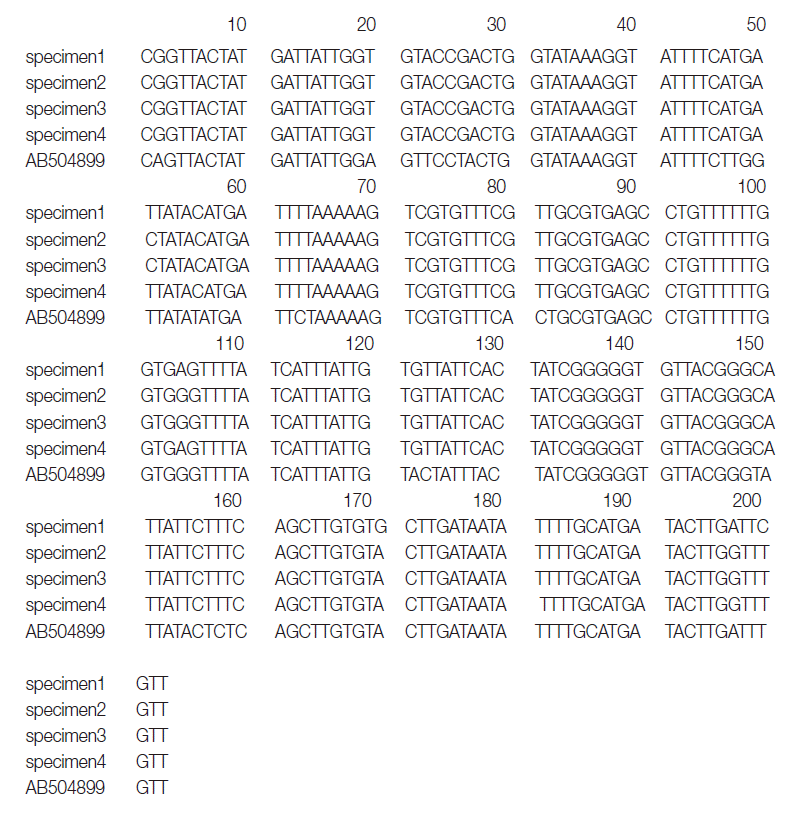
The primers used were D1f (5´-TGTGTGGGGGCATCATATGT-3´) and D1r (5´-ATGATAAGGAACAGGAGCTCAGCATA-3´). The second PCR and sequencing primers used were D1f1 (5´-GTTTAGATGTAAAGACGGCTG-3´) and D1r1 (5´-GGTAAACCGCACACACCAAA-3´) and yielded a 203-bp product corresponding to the position 919-1,122 bp of the
cox1 gene (
Fig. 1).
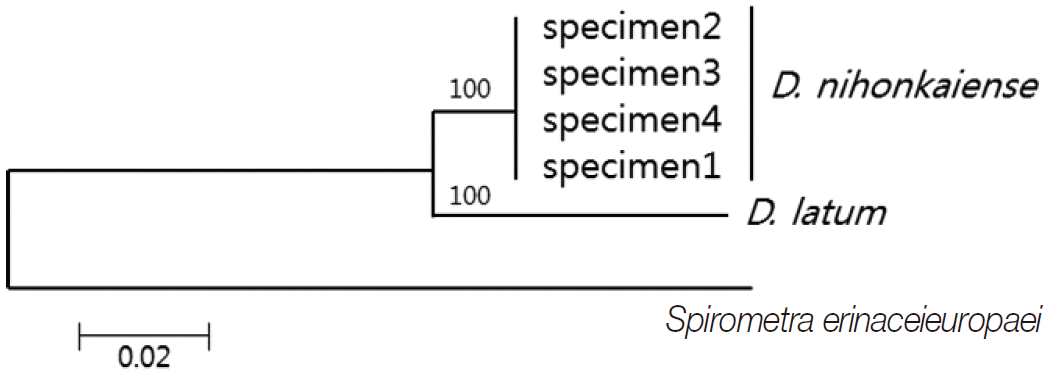
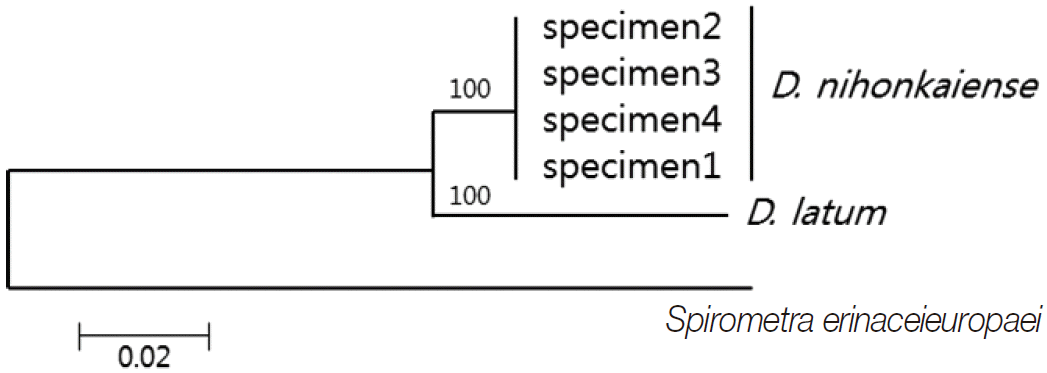
DNA sequencing was performed using a Big-Dye Terminator Sequencing kit (version 3.2, Applied Biosystems, Foster City, California, USA) and the reaction products were separated on electrophoresis using an automated DNA sequencer model 3739KL (Applied Biosystems). DNA sequences were assembled and aligned using Geneious 7.0.2 software (Biometer, Auckland, New Zealand). Phylogenetic analyses were performed using the neighbor-joining (NJ) method and the MEGA software (version 6). NJ analysis was performed using a distance matrix calculated with the Kimura 2 parameter method. The sequences were identified using BLAST searches and compared with sequences of
D. nihonkaiense and
D. latum deposited in the GenBank database. The
cox1 sequences (203 bp) of specimens 1, 2, 3, and 4 showed 99.5% (202/203), 98.5% (200/203), 98.5%, and 99.5% similarities to a reference
D. nihonkaiense sequence (NC 008945) and 91.6% (186/203) similarity to a reference
D. latum sequence (NC 008945), respectively (
Fig. 2).
DISCUSSION
D. nihonkaiense was recognized as a new species for the first time by Yamane et al. in 1986 [
11]. Jeon et al. [
11], Park et al. [
12], and Song et al. [
13] have identified
D. nihonkaiense in Korea. Before 2009,
D. latum has been primarily identified because of its morphological resemblance with
D. nihonkaiense, but after 2009,
D. nihonkaiense was confirmed by DNA analysis in all cases. Our additional 4 cases support the evidence that most or all of the diphyllobothriid tapeworms in Korea are
D. nihonkaiense but not
D. latum. In addition, there was no DNA evidence of
D. latum infection in Korea. In particular, patients with a salmon intake history are unlikely to be infected with
D. latum. Therefore, during an initial morphological diagnosis of
Diphyllobothrium infection,
D. nihonkaiense infection should be suspected first.
A single dose of praziquantel (10 mg/kg) has been recommended for the treatment of
D. nihonkaiense infection, similar to the dose used to treat
D. latum infection [
8]. However, in 1 case of suspected
D. nihonkaiense infection, a single dose of praziquantel caused a continuous stomachache, which required an additional dose for recovery [
14]. Therefore, 2 doses are thought to be necessary in some patients. Some authors have also used gastrograffin to treat
D. nihonkaiense infection [
15].
The limitation of this study was the small number of cases evaluated. Moreover, the medical history of 3 patients could not be obtained and evaluated. In conclusion, these 4 cases support the evidence of D. nihonkaiense infection in Korea. Further DNA sequencing studies are required for accumulation of evidence to prevent misidentification of D. latum in Korea.
Notes
-
We declare that we have no conflict of interest related to this work.
This research was supported by Prof. Sun Huh (Department of Parasitology, College of Medicine, Hallym University, Chuncheon, Korea) and we are grateful for his help.
Fig. 1.Aligned partial nucleotide sequences of a region of the mitochondrial cytochrome c oxidase subunit 1 gene of the DNA of adult Diphyllobothrium species. The sequences corresponded to 919-1,120 bp (total 203 bps) of the partial mitochondrial cox1 gene.
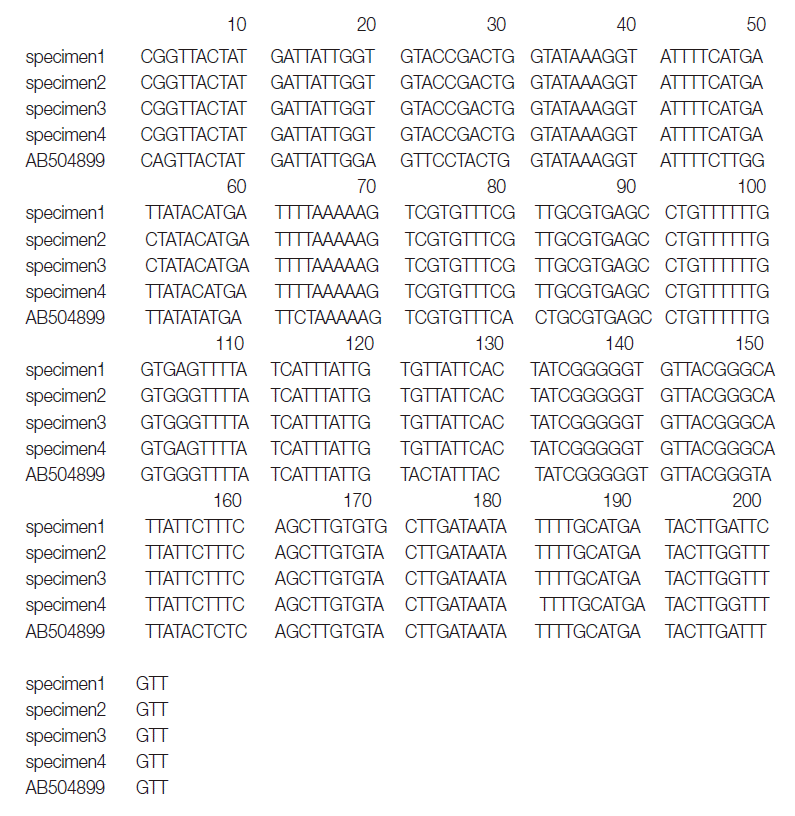
Fig. 2.Phylogenetic tree of Diphyllobothrium nihonkaiense and D. latum based on the partial cox1 sequences inferred from neighbor-joining (NJ) analyses. Numbers on branches indicate the bootstrap value based on 3,000 replicates. There were 203 base pairs corresponding to positions 919-1,122 bp of the cox1 gene.
References
- 1. Yanagida T, Matsuoka H, Kanai T, Nakao M, Ito A. Anomalous segmentation of Diphyllobothrium nihonkaiense. Parasitol Int 2010;59:268-270.
- 2. Chen S, Ai L, Zhang Y, Chen J, Zhang W, Li Y, Muto M, Morishima Y, Sugiyama H, Xu X, Zhou X, Yamasaki H. Molecular detection of Diphyllobothrium nihonkaiense in humans, China. Emerg Infect Dis 2014;20:315-318.
- 3. Yamasaki H, Kuramochi Y. A case of Diphyllobothrium nihonkaiense infection possibly linked to salmon consumption in New Zealand. Parasitol Res 2009;105:583-586.
- 4. Shimizu H, Kawakatsu H, Shimizu T, Yamada M, Tegoshi T, Uchikawa R, Arizono N. Diphyllobothriasis nihonkaiense: possibly acquired in Switzerland from imported Pacific salmon. Intern Med 2008;47:1359-1362.
- 5. Wicht B, Scholz T, Peduzzi R, Kuchta R. First record of human infection with the tapeworm Diphyllobothrium nihonkaiense in North America. Am J Trop Med Hyg 2008;78:235-238.
- 6. Yera H, Estran C, Delaunay P, Gari-Toussaint M, Dupouy-Camet J, Marty P. Putative Diphyllobothrium nihonkaiense acquired from a Pacific salmon (Oncorhynchus keta) eaten in France; genomic identification and case report. Parasitol Int 2006;55:45-49.
- 7. Wicht B, de Marval F, Peduzzi R. Diphyllobothrium nihonkaiense (Yamane et al., 1986) in Switzerland: first molecular evidence and case reports. Parasitol Int 2007;56:195-199.
- 8. Ohnishi K, Kato Y. Single low-dose treatment with praziquantel for Diphyllobothrium nihonkaiense infections. Intern Med 2003;42:41-43.
- 9. Choi HJ, Lee J, Yang HJ. Four human cases of Diphyllobothrium latum infection. Korean J Parasitol 2012;50:143-146.
- 10. Suzuki J, Murata R, Sadamasu K, Araki J. Detection and identification
of Diphyllobothrium nihonkaiense plerocercoids from wild Pacific salmon (Oncorhynchus spp.) in Japan. J Helminthol 2010;84:434-440.
- 11. Jeon HK, Kim KH, Huh S, Chai JY, Min DY, Rim HJ, Eom KS. Morphologic and genetic identification of Diphyllobothrium nihonkaiense in Korea. Korean J Parasitol 2009;47:369-375.
- 12. Park SH, Eom KS, Park MS, Kwon OK, Kim HS, Yoon JH. A case of Diphyllobothrium nihonkaiense infection as confirmed by mitochondrial COX1 gene sequence analysis. Korean J Parasitol 2013;51:471-473.
- 13. Song SM, Yang HW, Jung MK, Heo J, Cho CM, Goo YK, Hong Y, Chung DI. Two human cases of Diphyllobothrium nihonkaiense infection in Korea. Korean J Parasitol 2014;52:197-199.
- 14. Kim TH, Kim HK, Lee YS, Choi DH, Kang SH, Jeong SJ, Park TI, Kim IT. A case of Diphyllobothrium latum infection in a patient with abdominal pain. Korean J Gastroenterol 2007;50:384-387. (in Korean).
- 15. Ono S, Morimoto N, Korenaga M, Kumazawa H, Komatsu Y, Kuge I, Higashidani Y, Ogura K, Sugiura T. Molecular identification of human Diphyllobothrium nihonkaiense using mitochondrial cytochrome c oxidase subunit 1 (cox1) gene sequence. Rinsho Byori 2010;58:1085-1092. (in Japanese).