Abstract
Merozoite surface proteins (MSPs) of malaria parasites play critical roles during the erythrocyte invasion and so are potential candidates for malaria vaccine development. However, because MSPs are often under strong immune selection, they can exhibit extensive genetic diversity. The gene encoding the merozoite surface protein-3 (MSP-3) of Plasmodium falciparum displays 2 allelic types, K1 and 3D7. In Thailand, the allelic frequency of the P. falciparum msp-3 gene was evaluated in a single P. falciparum population in Tak at the Thailand and Myanmar border. However, no study has yet looked at the extent of genetic diversity of the msp-3 gene in P. falciparum populations in other localities. Here, we genotyped the msp-3 alleles of 63 P. falciparum samples collected from 5 geographical populations along the borders of Thailand with 3 neighboring countries (Myanmar, Laos, and Cambodia). Our study indicated that the K1 and 3D7 alleles coexisted, but at different proportions in different Thai P. falciparum populations. K1 was more prevalent in populations at the Thailand-Myanmar and Thailand-Cambodia borders, whilst 3D7 was more prevalent at the Thailand-Laos border. Global analysis of the msp-3 allele frequencies revealed that proportions of K1 and 3D7 alleles of msp-3 also varied in different continents, suggesting the divergence of malaria parasite populations. In conclusion, the variation in the msp-3 allelic patterns of P. falciparum in Thailand provides fundamental knowledge for inferring the P. falciparum population structure and for the best design of msp-3 based malaria vaccines.
-
Key words: Plasmodium falciparum, genotyping, merozoite surface protein-3, molecular epidemiology
INTRODUCTION
Human malaria is caused by parasitic protozoa in the genus
Plasmodium. Of the 5 species of malaria parasites that infect humans,
Plasmodium falciparum is the most deadly and is responsible for severe and lethal cases, thereby causing a major public health concern [
1]. Around 2-3 billion people in the tropics and subtropics live in malaria endemic regions and are at risk of exposure to malarial infections. Each year, there are 200 million actual illnesses from malaria and up to 0.6-1 million deaths [
2]. With the rapid growth of human populations in regions with high malaria transmission, it has been estimated that, in the absence of effective interventions, the number of malaria cases will double by 2021 [
3]. Within the Southeast Asia region, Thailand and its neighboring countries, such as Myanmar and Cambodia, are major hot spots for anti-malarial drug resistance [
4]. The first incidence of artemisinin resistance in
P. falciparum was reported on the Thailand-Cambodian border in 2008 [
5], and the resistance later emerged on the western border of Thailand close to Myanmar and so is now likely to spread to other regions [
6,
7]. This highlights the importance of deploying effective control measures to delay or prevent the spread of the parasite.
Because the pathology and clinical manifestations of malaria are mainly attributed to the erythrocytic stage development of the
Plasmodium parasite’s life cycle, then vaccines against the erythrocytic stage have been developed to induce protective immunity against these parasites [
8,
9]. This type of vaccine could prevent invasion of the merozoite into erythrocytes, inhibit the development of the parasite in erythrocytes or speed up the clearance of parasitized erythrocyte. The most common blood stage vaccine candidates are the membrane proteins of the merozoites such as merozoite surface protein-1 (MSP-1), merozoite surface protein-2 (MSP-2), and apical membrane antigen-1 (AMA-1) [
10]. While these antigens have been extensively studied, there are many other membrane proteins such as merozoite surface protein-3 (MSP-3) that have received much less attention, but could be considered potential candidates for blood stage vaccine development.
MSP-3, also known as secreted polymorphic antigen associated with merozoites (SPAM), is one of the essential proteins associated with the merozoite surface, although it lacks a hydrophobic transmembrane domain or a GPI anchor [
11]. MSP-3 was first identified using human hyperimmune serum with monocytes in an antibody-dependent cellular inhibition to screen for antibodies that inhibited the
P. falciparum erythrocytic stage growth in vitro [
12,
13]. The antigen responsible was found to be MSP-3, suggesting its involvement in the invasion of erythrocytes. Immunizations with full-length or truncated forms of
P. falciparum MSP-3 also elicited full or partial protection from a challenge infection with
P. falciparum in a Saimiri model [
14]. Furthermore, evidence from epidemiological and immunological studies showed that the levels of IgG3 antibodies against the allele-specific and conserved epitopes in MSP-3 were strongly associated with protection from clinical malaria [
15-
18]. Peptides derived from MSP-3 protein of
P. falciparum have now been incorporated and tested as erythrocytic stage malaria vaccines in field trials [
19-
22].
P. falciparum MSP-3 is a 48 kDa protein and is composed of 3 blocks of (i) 4 heptad repeats at the N-termial with the hydrophobic amino acid alanine (A) in the 1st and 4th positions (AXXAXXX motif) of each haptad, (ii) a hydrophobic glutamine rich domain, and (iii) a putative leucine zipper domain at the C-terminal [
23]. It is encoded by a single-copy
msp-3 gene on chromosome 10 (PF10_0345). The
msp-3 gene exists as the 2 allelic types, K1 and 3D7, that differ in the size variation of the heptad repeat region at the N-terminal [
24].
Despite its immunological significance, there is currently very limited data on the extent of the genetic diversity of
P. falciparum msp-3 in natural populations. To date, only a few studies have evaluated the natural variation in the
msp-3 sequences of
P. falciparum field isolates. Analysis of
P. falciparum samples collected from Peru between 2003 and 2006 revealed the temporal variation of 3D7 and K1 alleles over the 4-year period [
25]. The 3D7 and K1 alleles of
msp-3 also appeared to be in equal proportion in parasite populations in Iran [
26]. Additionally, the analyses of
msp-3 allele frequencies in
P. falciparum samples collected from 6 countries in Central and West Africa between 2007 and 2009 showed a high degree of genetic homogeneity between the countries [
27]. Genotyping of
msp-3 from 48 and 50
P. falciparum samples from Nigeria and Thailand revealed that the polymorphism of
msp-3 was likely to be maintained under frequency-dependent selection [
28]. However, in the latter study, the
msp-3 sequence data from Thailand was derived from only a single population at Tak Province on the Thailand-Myanmar border and, to date, there have been no further reports that elucidate the extent of genetic diversity in other regions of the country or other countries in Southeast Asia.
Here, a cross-sectional survey of the allelic types and distribution patterns of the msp-3 gene in 5 geographical populations of P. falciparum in Thailand, located along the national borders of Thailand and 3 neighboring countries (Myanmar, Cambodia, and Laos), was performed on samples collected between 2002 and 2010. This data will provide a new insight into the genetic structure of natural populations of P. falciparum in Thailand and also generate the baseline epidemiological data for further studies on field trials of MSP-3 based vaccines.
MATERIALS AND METHODS
Study locations and parasite collections
A total of 63 blood samples, each from a patient infected with a single
P. falciparum strain (mono-infection), were collected from patients enrolled in each of the 5 sampling localities between 2002 and 2010 (
Table 1). The study sites were Mae Hong Son, Kanchanaburi, and Ranong at the Thailand-Myanmar border. The other study sites were Ubon Ratchathani and Trat at the Thai-Laos and the Thai-Cambodia borders, respectively (
Fig. 1). A detailed description of the study locations has been previously described [
28]. The procedures of parasite collections and maintenance were performed as previously described [
28]. The parasite species were confirmed by microscopic examinations of Giemsa-stained thin blood smears and verified by PCR using primers specific to the
P. falciparum cytochrome B gene (data not shown). Forty samples collected between 2002 and 2006 were previously genotyped by 12 microsatellite loci and shown to be independent clones [
29]. The parasites were grown to a parasitemia level of 5-10% and harvested for genomic DNA preparation. The origins of
P. falciparum reference clones 3D7 and K1CB1 (K1) were described previously [
28].
The standard phenol/chloroform DNA extraction method was used to extract genomic DNA from the infected blood. In brief, a total of 200 μl of the packed blood cells was mixed with 0.05% (w/v) saponin solution in PBS (pH 7.4). The parasite pellets were mixed with 400 μl of lysis solution (40 mM Tris-HCl, 80 mM EDTA, 2% w/v sodium dodecyl sulfate, pH 8.0) containing 2 mg/ml proteinase K and incubated at 42˚C overnight. The aqueous phase was sequentially extracted with an equal volume of Tris-HCl saturated phenol (pH 8.0), phenol/chloroform/isoamyl alcohol (25:24:1 v/v/v, pH 8.0) and chloroform, harvesting the aqueous phase each time. The DNA was precipitated from the aqueous phase with the addition of a 0.1× volume of 0.3 M sodium acetate (pH 5.2) and a 1× volume of absolute ethanol and centrifugation. The genomic DNA pellets were washed with 70% (v/v) ethanol and later resuspended in standard TE buffer (10 mM Tris-HCl, 0.1 mM Na2EDTA, pH 8.0) and stored at -20˚C prior to PCR genotyping.
Genotyping of the P. falciparum merozoite surface protein-3 gene
Primers for genotyping of the
msp-3 gene (PF10_ 0345) were M3F/O 5´-ATGAAAAGTTTTAT AAATATTACTCTTTC-3´ and M3R/O 5´-CATGTTATGAATATAAATTATGTTCA-3, which correspond to nucleotide positions 1404192-1404220 (the start codon at positions 1404192-1404194) and 1405293-1405268 (the stop codon at positions 1405254-1405256) of the chromosome 10 of
P. falciparum strain 3D7 (NCBI accession no.AE014185.2) [
30]. The standard PCR reaction was performed in a total volume of 50 μl, containing 200-300 ng of DNA templates, 2 mM of MgCl
2, 200 μM of dNTPs, 0.5 μM of each primer and 2 units of FastStart
Taq DNA polymerase enzyme in 1×
Taq PCR buffer (Roche Diagnostics, Mannheim, Germany). Thermal cycling was performed with an optimized profile of an initial denaturation at 95˚C for 5 min, followed by 40 cycles of 95˚C for 40 sec, 56˚C for 40 sec, 68˚C for 80 sec, and then followed by a final extension at 68˚C for 10 min. Subsequently, PCR products were analyzed on 2% (w/v) agarose gel. Electrophoresis condition was at 80 V for 40 min in 1× TBE buffer. PCR products were visualized and photographed under UV transillumination after ethidium bromide staining. The genotypes of the strains 3D7 and K1 of
P. falciparum were used as the reference samples (and positive controls). Each sample was genotyped at least 3 times. The allelic types were further confirmed by DNA sequencing of the N-terminal coding region (data not shown).
The data was expressed as the frequencies of the 3D7 and K1 alleles in each sampled parasite population, where any samples with a mixed genotype were first excluded from the analyses. Population differentiation of the
msp-3 allele frequencies between 2 parasite populations was tested using Wright’s fixation index (
Fst) in Arlequin suite version 3.5 [
31]. Statistical differences were accepted at
P<0.05.
RESULTS
Genotypes and distribution patterns of the msp-3 gene in Thai P. falciparum populations
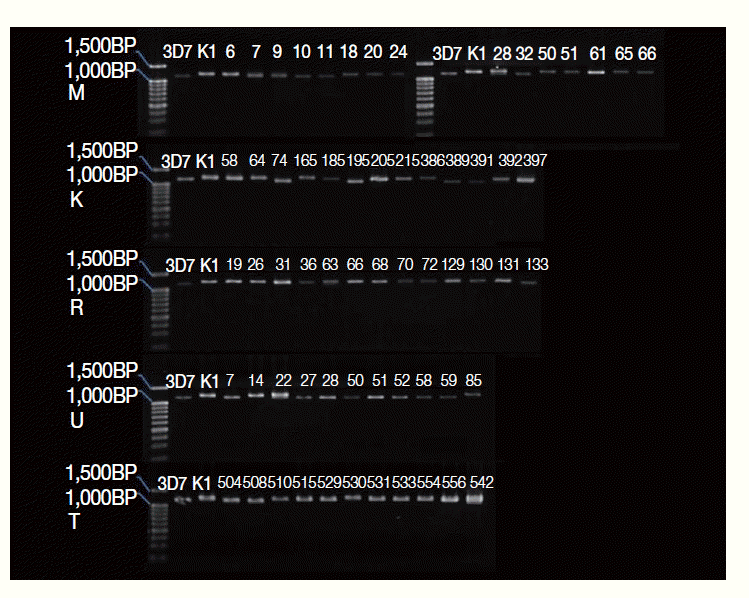
Genotypes of the
msp-3 alleles of 63
P. falciparum isolates in Thailand were analyzed by PCR and standard agarose gel electrophoresis as described in Materials and Methods. The genotyping results showed that 89% (56 samples) of the blood samples were comprised of single
msp-3 allelic types of either 3D7 or K1, while 11% (7 samples) had mixed
msp-3 allelic types (
Figs. 1,
2). The mixed
msp-3 genotypes were detected in 20%, 15%, 9%, and 9% of
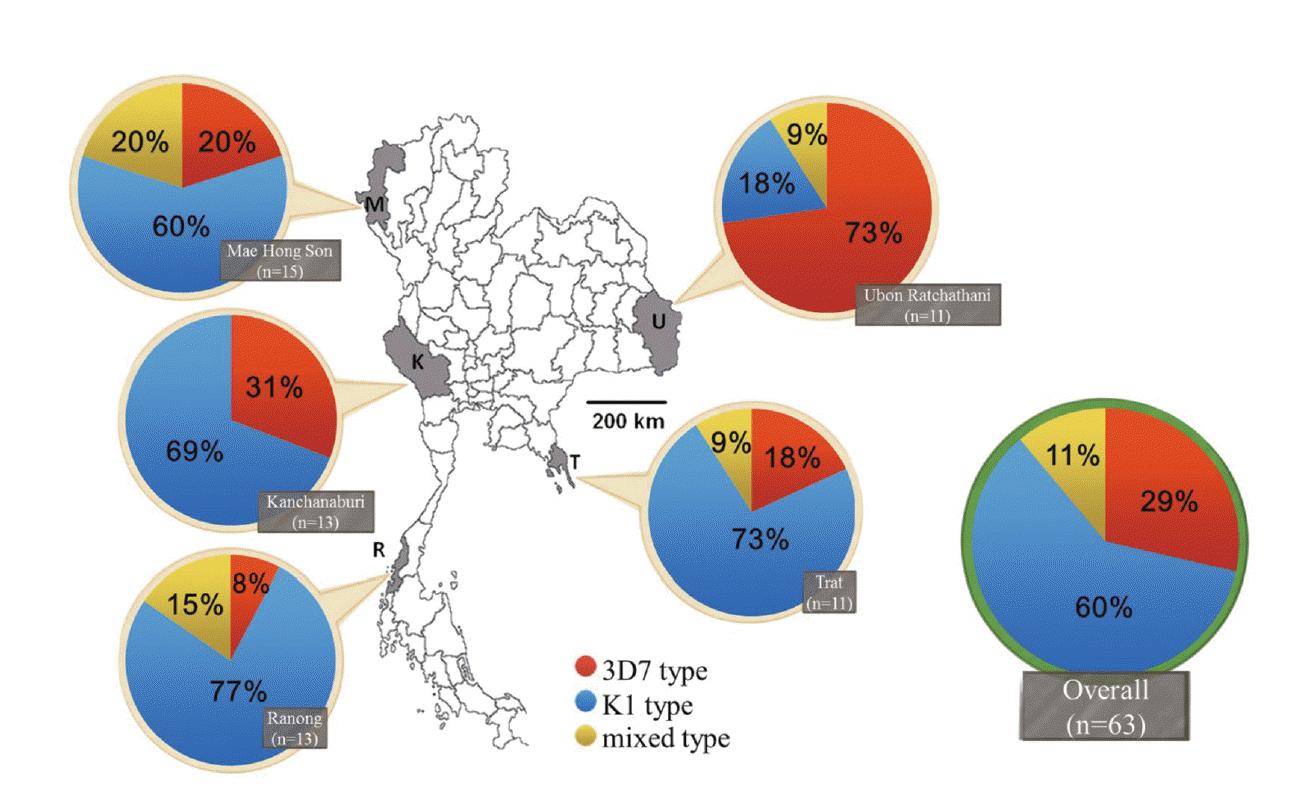
P. falciparum samples in Mae Hong Son, Ranong, Ubon Ratchatani, and Trat, respectively.
Of the 56 samples with a single
msp-3 allelic type, 29% (18 samples) and 60% (38 samples) were unambiguously classified as 3D7 and K1 subtypes, respectively (
Fig. 1). The K1 subtype was highly prevalent in 4
P. falciparum populations in Trat, Mae Hong Son, Kanchanaburi, and Ranong, representing 73%, 60%, 69%, and 77% of the populations. In contrast, the 3D7 subtype was more prevalent in
P. falciparum populations in Ubon Ratchatani, representing 73% of the population. Thus, the 3D7 and K1 allelic types of
msp-3 circulated in
P. falciparum Thai populations, but the allele frequency varied between the different geographic populations.
To further determine whether the parasite populations at the borders of Thailand and the 3 neighboring countries were genetically homogenous, pairwise inter-population comparisons were performed for each parasite population using Wright’s fixation index (
Fst). In this analysis, allele frequency of
msp-3 from a
P. falciparum population in Tak Province (10 and 40 samples collected in 2000 with single 3D7 and K1 types, respectively), which is located between Mae Hong Son and Kanchanaburi, was also included.
Table 2 shows the
Fst values from the pairs of all 6 parasite populations, where the
P. falciparum population in Ubon Ratchatani appeared to be genetically distinct from those in the other malaria endemic regions in Thailand (
P<0.05). These results suggested that the
P. falciparum populations in Thailand could be divided according to the
msp-3 alleles into 2 subpopulations. The major group, with the predominance of K1 allele, was comprised of populations at the Thailand-Myanmar border (Mae Hong Son, Tak, Kanchanburi, and Ranong) and also the population at the Thailand-Cambodia border in Trat. The other minor population, in which the 3D7
msp-3 allele was predominant, was the parasite population in Ubon Ratchatani at the Thai-Laos border.
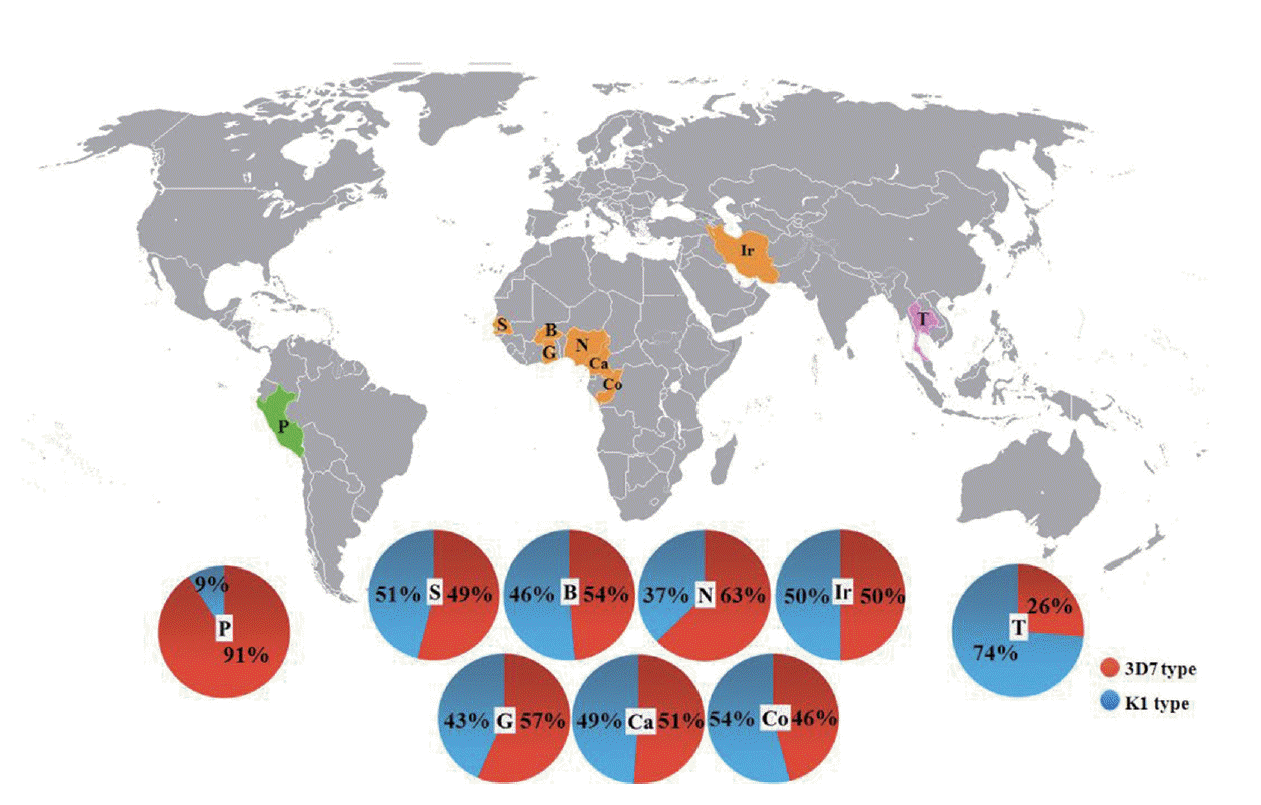
The allele frequencies of
msp-3 from 6
P. falciparum populations in Africa (Nigeria, Republic of Congo, Cameroon, Ghana, Burkina Faso, and Senegal), 1 parasite population in the Middle East (Iran), and 1 parasite population in South America (Peru) were obtained through the literature search at the NCBI and Scopus databases (
Fig. 3;
Tables 3,
4). While the K1 allele was the dominant allelic type in Thailand (data inclusive of the genotypes in Tak [
17]), the 3D7 allele was the dominant allelic type in South America, and the K1 and 3D7 allele proportions were close to 1:1 in most Central and West African parasite populations and in Iranian population.
Pair-wise inter-population comparisons, performed for each of the parasite populations using the Wright’s fixation index revealed that the
Fst values were low and non-significant (
P>0.05) between the pairs of the 6 parasite populations within Africa (
Table 4). Likewise, low and non-significant
Fst values (
P>0.05) were found when comparing the
msp-3 allele frequency between the parasite populations from Iran and African countries (
Table 4). In contrast, significant
Fst values (
P<0.05) were detected between parasite populations from Thailand and Peru, Thailand and Iran, Iran and Peru, Thailand and the 6 African countries, and between Peru and the 6 African countries. Thus, the overall allele patterns of
msp-3 parasite populations in Thailand, Peru, and the 6 African countries (plus Iran) were different, suggesting that the populations of
P. falciparum parasites in 3 continents were geographically and genetically isolated or subject to different allele-specific selection processes.
DISCUSSION
The nature and extent of genetic diversity within and between populations of malaria parasites is essential knowledge for understanding the mechanisms underlying the phenotypic variation and in inferring their population structures. The genetic diversity of the human malaria parasites in Southeast Asia has been a subject of intensive research, which is partly because Thailand and the neighboring countries are major hot spots of drug resistance [
4,
32]. Several polymorphic markers have been developed and employed for the genotyping of the malaria parasites. These include microsatellite markers and the genes encoding surface antigens such as
msp-1,
msp-2, and
msp-3 [
29,
33,
34]. However, in the case of
msp-3 for
P. falciparum in Thailand, allelic diversity has only been surveyed in a single population in Tak Province, located at the Thailand-Myanmar border [
17]. Therefore, the aim of this work was to determine the allelic frequency and distribution patterns for the
msp-3 gene in 5 geographical regions of Thailand.
Both K1 and 3D7 alleles of
msp-3 were found to co-circulate, but the proportions of individual populations varied according to geographical parasite populations. The K1-type of
msp-3 was more prevalent in 3 parasite populations in Mae Hong Son, Kanchanaburi, and Ranong, located along the Thailand and Myanmar borders. This finding was in agreement with the previous observations in Tak [
17], indicating the dominance of K1-allele of
msp-3 in the western population of Thailand. Likewise, the dominance of K-1 allele was also observed in a parasite population in Trat, located at the border of Thailand and Cambodia. The similar genetic patterns of the parasite between these 2 regions were detected using polymorphic marker
msp-3α and -3β of
Plasmodium vivax [
35]. These findings suggest evidence of gene flow between the parasite populations that is due likely to the population movement. In contrast, the dominance of 3D7 allele was detected in a parasite population in Ubon Ratchatani. In this study, the genetic differentiation of
P. falciparum parasites in Thailand was unambiguously divided into 2 sub-populations. The first was those from the western border of Thailand-Myanmar and also at the Thailand-Cambodia border. The second was a minor population, represented by the population at the Thailand-Laos border. Overall, these results were in agreement with the observations using neutral, genome-wide microsatellite markers that demonstrated the population differentiations between the populations at the eastern and western borders of Thailand [
29]. Also, noteworthy is that the population structure based on
msp-3 and microsatellite typing was contradictory to that based on the genotypes of the msp-1 gene block 17, which was previously demonstrated that the 5 geographical parasite populations in Thailand were genetically homogenous [
28]. This discrepancy may be due to the msp-1 gene block 17 being under functional constraints that limited its genetic polymorphism [
36,
37], or to the relatively small number of samples analyzed.
Extensive human migration in the border areas may have also led to the introduction of new genotypes, resulting in the increase in the genetic diversity [
38,
39]. A collection of a larger number of samples should be considered for analysis in future studies. Inclusions of
P. falciparum samples from Myanmar, Laos, and Cambodia would also be of great value to better understand the true epidemiological structure of the malaria parasites in the Indochina region.
In addition, we also conducted population differentiation analysis and included the
msp-3 genotype data from 8 distinct parasite populations in African countries, Iran, and Peru [
17,
25-
27]. This analysis demonstrated the differences in the allele frequency and patterns of
msp-3 among the parasite populations in different continents. Using the population differentiation statistics, our study suggested that the population structure of
P. falciparum in Thailand, Africa, and South America were distinct from each other, thereby suggesting that the gene flow between the parasites in these regions was highly unlikely. Our analysis also showed the similar allelic pattern of
msp-3 between parasite populations from Iran and Africa, suggesting the close genetic relationships or gene flow between these populations. Regardless, this information is very crucial for the design of MSP-3 based vaccine. If the full-length MSP-3 molecule was to be developed as a vaccine, it would be preferable to incorporate both the K1 and 3D7 alleles that match the
msp-3 allele frequency for each parasite population, as such a multivalent vaccine would have more long-term usefulness for induction of protective immunity [
40].
This study extends our current understanding and knowledge of the variation and prevalence of the msp-3 alleles in natural populations of P. falciparum in Thailand. We showed that the K1 allele was the major variant of the msp-3 gene in P. falciparum populations. The msp-3 allele frequency varied between different geographical locations, where gene flow may occur in P. falciparum populations in Thailand and neighboring countries, but not between other continents. Finally, the present findings provide an overview of the population structure and dynamics of the malaria parasite that is critical for monitoring the population responses to MSP-3 based vaccines in clinical trials.
Notes
-
We have no conflict of interest related to this work.
We would like to thank Associate Professor Dr. Chanpen Chanchao (Department of Biology, Faculty of Science, Chulalongkorn University, Thailand) for sharing laboratory equipment and Dr. Robert Douglas John Butcher (Publication Counseling Unit, Faculty of Science, Chulalongkorn University) for English language editing. This work was supported by a grant from the Thailand Research Fund (TRF) (MRG5680134), a grant for Development of New Faculty Staff, Chulalongkorn University, Bangkok, Thailand (GDNS 56-064-23-017 and GDNS-58-026-23-005) to Sittiporn Pattaradilokrat and a grant from Development and Promotion of Science and Technology (DPST) Project, the Royal Thai Government to Vorthon Sawaswong. The funders had no role in study design, data collection and analysis, decision to publish, and preparation of the manuscript.
Fig. 1.The distribution of the alleles of merozoites surface protein-3 gene in 5 geographical populations of P. falciparum in Thailand. The 5 sampling locations were Mae Hong Son (M), Kanchanaburi (K), and Ranong (R), located at the Thailand-Myanmar border, Ubon Ratchathani (U) located at the Thailand-Laos border, and Trat (T) located at the Thailand-Cambodia border. Numbers (n) in the grey boxes indicate the numbers of parasites from each sampling site or the total number of parasite isolates in Thailand. Numbers in the pie charts represent the percentage of the 3D7-type (red), K1-type (blue) or mixed-type (yellow).

Fig. 2.Genotypes of the merozoite surface protein-3 gene of P. falciparum populations in Thailand. Genomic DNA samples obtained from P. falciparum parasites in 5 geographical locations, Mae Hong Son (M, n=15), Kanchanaburi (K, n=13), Ranong (R, n=13), Ubon Ratchantani (U, n=11), and Trat (T, n=11), were PCR amplified, resolved, and visualized as described in Materials and Methods. The alleles of the msp-3 gene were classified into the 3D7, K1, or mixed type.

Fig. 3.Global distribution of the merozoite surface protein-3 alleles of P. falciparum. Numbers in pie charts represent percentages of the 3D7 (red) and K1 (blue) alleles. Data of the Thai population (T) included the genotyping data from P. falciparum population in Tak. Countries from which msp-3 genotypes were available were: Iran (Ir), Peru (P), Senegal (S), Ghana (G), Burkina Faso (B), Cameroon, (Ca), Republic of Congo (Co), and Nigeria (N).

Table 1.The sample location, year of collection, and genotype of the merozoite surface protein-3 gene of Plasmodium falciparum
Table 1.
|
Origin of Pf isolate |
Name of isolate |
Year of collection |
Genotype of the msp-3 gene |
|
Origin of Pf isolate |
Name of isolate |
Year of collection |
Genotype of the msp-3 gene |
|
Kanchanaburi |
K165*
|
2005 |
K1 |
|
Ranong |
RN19*
|
2003 |
K1 |
|
Kanchanaburi |
K185*
|
2005 |
K1 |
|
Ranong |
RN26*
|
2003 |
K1 |
|
Kanchanaburi |
K195*
|
2005 |
3D7 |
|
Ranong |
RN31*
|
2003 |
K1 |
|
Kanchanaburi |
K205*
|
2005 |
K1 |
|
Ranong |
RN36*
|
2003 |
K1 |
|
Kanchanaburi |
K215*
|
2005 |
K1 |
|
Ranong |
RN63*
|
2005 |
Mixed |
|
Kanchanaburi |
K386 |
2008 |
K1 |
|
Ranong |
RN66*
|
2005 |
K1 |
|
Kanchanaburi |
K389 |
2008 |
3D7 |
|
Ranong |
RN68*
|
2005 |
K1 |
|
Kanchanaburi |
K391 |
2008 |
3D7 |
|
Ranong |
RN70*
|
2005 |
Mixed |
|
Kanchanaburi |
K392 |
2008 |
K1 |
|
Ranong |
RN72*
|
2005 |
K1 |
|
Kanchanaburi |
K397 |
2008 |
K1 |
|
Trat |
TD504 |
2003 |
3D7 |
|
Kanchanaburi |
K58*
|
2002 |
K1 |
|
Trat |
TD508*
|
2003 |
3D7 |
|
Kanchanaburi |
K64*
|
2002 |
K1 |
|
Trat |
TD510*
|
2003 |
K1 |
|
Kanchanaburi |
K74*
|
2002 |
3D7 |
|
Trat |
TD515*
|
2003 |
K1 |
|
Mae Hong Son |
MH06 |
2003 |
K1 |
|
Trat |
TD529*
|
2005 |
K1 |
|
Mae Hong Son |
MH07 |
2003 |
Mixed |
|
Trat |
TD530*
|
2005 |
K1 |
|
Mae Hong Son |
MH09*
|
2003 |
Mixed |
|
Trat |
TD531*
|
2005 |
K1 |
|
Mae Hong Son |
MH10*
|
2003 |
3D7 |
|
Trat |
TD533 |
2005 |
K1 |
|
Mae Hong Son |
MH11*
|
2004 |
3D7 |
|
Trat |
TD542*
|
2006 |
Mixed |
|
Mae Hong Son |
MH18*
|
2005 |
K1 |
|
Trat |
TD554 |
2008 |
K1 |
|
Mae Hong Son |
MH20*
|
2005 |
K1 |
|
Trat |
TD556 |
2008 |
K1 |
|
Mae Hong Son |
MH24*
|
2005 |
K1 |
|
Ubon Ratchathani |
UB14*
|
2003 |
K1 |
|
Mae Hong Son |
MH28*
|
2005 |
Mixed |
|
Ubon Ratchathani |
UB22*
|
2003 |
Mixed |
|
Mae Hong Son |
MH32*
|
2005 |
3D7 |
|
Ubon Ratchathani |
UB27*
|
2003 |
3D7 |
|
Mae Hong Son |
MH50 |
2010 |
K1 |
|
Ubon Ratchathani |
UB28*
|
2003 |
3D7 |
|
Mae Hong Son |
MH51 |
2010 |
K1 |
|
Ubon Ratchathani |
UB50*
|
2005 |
3D7 |
|
Mae Hong Son |
MH61 |
2010 |
K1 |
|
Ubon Ratchathani |
UB51*
|
2005 |
3D7 |
|
Mae Hong Son |
MH65 |
2010 |
K1 |
|
Ubon Ratchathani |
UB52 |
2005 |
3D7 |
|
Mae Hong Son |
MH66 |
2010 |
K1 |
|
Ubon Ratchathani |
UB58 |
2005 |
3D7 |
|
Ranong |
RN129 |
2008 |
K1 |
|
Ubon Ratchathani |
UB59*
|
2005 |
3D7 |
|
Ranong |
RN130 |
2008 |
K1 |
|
Ubon Ratchathani |
UB7*
|
2003 |
3D7 |
|
Ranong |
RN131 |
2008 |
K1 |
|
Ubon Ratchathani |
UB85 |
2008 |
K1 |
|
Ranong |
RN133 |
2008 |
3D7 |
|
|
|
|
|
Table 2.Pairwise Fst values of the msp-3 alleles in P. falciparum populations in Thailand
Table 2.
|
Mae Hong Son |
Ubon Ratchathani |
Kanchanaburi |
Trat |
Ranong |
|
Ubon Ratchathani |
0.40970* (P=0.03) |
- |
|
|
|
|
Kanchanaburi |
-0.07817 (P=0.99) |
0.33065* (P=0.04) |
- |
|
|
|
Trat |
-0.09310 (P=0.99) |
0.47712* (P=0.03) |
-0.06450 (P=0.65) |
- |
|
|
Ranong |
-0.00301 (P=0.56) |
0.64391** (P=0.00) |
0.05359 (P=0.33) |
-0.05307 (P=0.62) |
- |
|
Taka
|
-0.04641 (P=0.99) |
0.50678** (P=0.00) |
-0.01586 (P=0.47) |
-0.06383 (P=0.99) |
-0.01631 (P=0.71) |
Table 3.The number of 3D7-type and K1-type alleles of the merozoite surface protein-3 gene in the Plasmodium falciparum populations in Thailand, 6 African countries, Iran (the Middle East) and Peru (South America)
Table 3.
|
Country |
Total |
3D7 type |
K1 type |
References |
|
Thailand |
106 |
28 |
78 |
Data of the present work and that of [17] |
|
Nigeria |
51 |
32 |
19 |
[17] |
|
Republic of Congo |
85 |
39 |
46 |
[27] |
|
Cameroon |
90 |
46 |
44 |
[27] |
|
Ghana |
83 |
47 |
36 |
[27] |
|
Burkina Faso |
216 |
105 |
111 |
[27] |
|
Senegal |
98 |
53 |
45 |
[27] |
|
Peru |
627 |
570 |
57 |
[25] |
|
Total |
1,356 |
920 |
436 |
|
Table 4.Pairwise Fst values of the msp-3 alleles in P. falciparum populations in Thailand and other malaria endemic regions
Table 4.
|
P. falciparum populations |
Thailanda
|
Nigeriab
|
Republic of Congob
|
Cameroonb
|
Ghanab
|
Burkina Fasob
|
Senegalb
|
Iranb
|
|
Nigeria |
0.23075** (P=0.00) |
|
|
|
|
|
|
|
|
Republic of Congo |
0.07000** (P=0.01) |
0.04031 (P=0.07) |
|
|
|
|
|
|
|
Cameroon |
0.11270** (P=0.00) |
0.01184 (P=0.22) |
-0.00605 (P=0.55) |
|
|
|
|
|
|
Ghana |
0.16490** (P=0.00) |
-0.00825 (P=0.60) |
0.01106 (P=0.17) |
-0.00554 (P=0.54) |
|
|
|
|
|
Burkina Faso |
0.08982** (P=0.00) |
0.02729 (P=0.09) |
-0.00676 (P=0.69) |
-0.00667 (P=0.71) |
0.00447 (P=0.26) |
|
|
|
|
Senegal |
0.13957** (P=0.00) |
0.00031 (P=0.40) |
0.0024 (P=0.30) |
-0.00899 (P=0.77) |
-0.00993 (P=0.76) |
-0.00146 (P=0.40) |
|
|
|
Iran |
0.10221** (P=0.00) |
0.01888 (P=0.12) |
-0.00623 (P=0.62) |
-0.00907 (P=0.92) |
-0.00094 (P=0.36) |
-0.000256 (P=0.80) |
-0.0055 (P=0.62) |
|
|
Peru |
0.67676** (P=0.00) |
0.29079** (P=0.00) |
0.49502** (P=0.00) |
0.43042** (P=0.00) |
0.36281** (P=0.00) |
0.41456** (P=0.00) |
0.38946** (P=0.00) |
0.42448** (P=0.00) |
References
- 1. Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature 2002;415:673-679.
- 2. Murray CJ, Ortblad KF, Guinovart C, Lim SS, Wolock TM, Roberts DA, Dansereau EA, Graetz N, Barber RM, Brown JC, Wang H, Duber HC, Naghavi M, Dicker D, Dandona L, Salomon JA, Heuton KR, Foreman K, Phillips DE, Fleming TD, Flaxman AD, Phillips BK, Johnson EK, Coggeshall MS, Abd-Allah F, Abera SF, Abraham JP, Abubakar I, Abu-Raddad LJ, Abu-Rmeileh NM, Achoki T, Adeyemo AO, Adou AK, Adsuar JC, Agardh EE, Akena D, Al Kahbouri MJ, Alasfoor D, Albittar MI, Alcalá-Cerra G, Alegretti MA, Alemu ZA, Alfonso-Cristancho R, Alhabib S, Ali R, Alla F, Allen PJ, Alsharif U, Alvarez E, Alvis-Guzman N, Amankwaa AA, Amare AT, Amini H, Ammar W, Anderson BO, Antonio CA, Anwari P, Arnlöv J, Arsenijevic VS, Artaman A, Asghar RJ, Assadi R, Atkins LS, Badawi A, Balakrishnan K, Banerjee A, Basu S, Beardsley J, Bekele T, Bell ML, Bernabe E, Beyene TJ, Bhala N, Bhalla A, Bhutta ZA, Abdulhak AB, Binagwaho A, Blore JD, Basara BB, Bose D, Brainin M, Breitborde N, Castañeda-Orjuela CA, Catalá-López F, Chadha VK, Chang JC, Chiang PP, Chuang TW, Colomar M, Cooper LT, Cooper C, Courville KJ, Cowie BC, Criqui MH, Dandona R, Dayama A, De Leo D, Degenhardt L, Del Pozo-Cruz B, Deribe K, Des Jarlais DC, Dessalegn M, Dharmaratne SD, Dilmen U, Ding EL, Driscoll TR, Durrani AM, Ellenbogen RG, Ermakov SP, Esteghamati A, Faraon EJ, Farzadfar F, Fereshtehnejad SM, Fijabi DO, Forouzanfar MH, Fra Paleo U, Gaffikin L, Gamkrelidze A, Gankpé FG, Geleijnse JM, Gessner BD, Gibney KB, Ginawi IA, Glaser EL, Gona P, Goto A, Gouda HN, Gugnani HC, Gupta R, Gupta R, Hafezi-Nejad N, Hamadeh RR, Hammami M, Hankey GJ, Harb HL, Haro JM, Havmoeller R, Hay SI, Hedayati MT, Pi IB, Hoek HW, Hornberger JC, Hosgood HD, Hotez PJ, Hoy DG, Huang JJ, Iburg KM, Idrisov BT, Innos K, Jacobsen KH, Jeemon P, Jensen PN, Jha V, Jiang G, Jonas JB, Juel K, Kan H, Kankindi I, Karam NE, Karch A, Karema CK, Kaul A, Kawakami N, Kazi DS, Kemp AH, Kengne AP, Keren A, Kereselidze M, Khader YS, Khalifa SE, Khan EA, Khang YH, Khonelidze I, Kinfu Y, Kinge JM, Knibbs L, Kokubo Y, Kosen S, Defo BK, Kulkarni VS, Kulkarni C, Kumar K, Kumar RB, Kumar GA, Kwan GF, Lai T, Balaji AL, Lam H, Lan Q, Lansingh VC, Larson HJ, Larsson A, Lee JT, Leigh J, Leinsalu M, Leung R, Li Y, Li Y, De Lima GM, Lin HH, Lipshultz SE, Liu S, Liu Y, Lloyd BK, Lotufo PA, Machado VM, Maclachlan JH, Magis-Rodriguez C, Majdan M, Mapoma CC, Marcenes W, Marzan MB, Masci JR, Mashal MT, Mason-Jones AJ, Mayosi BM, Mazorodze TT, Mckay AC, Meaney PA, Mehndiratta MM, Mejia-Rodriguez F, Melaku YA, Memish ZA, Mendoza W, Miller TR, Mills EJ, Mohammad KA, Mokdad AH, Mola GL, Monasta L, Montico M, Moore AR, Mori R, Moturi WN, Mukaigawara M, Murthy KS, Naheed A, Naidoo KS, Naldi L, Nangia V, Narayan KM, Nash D, Nejjari C, Nelson RG, Neupane SP, Newton CR, Ng M, Nisar MI, Nolte S, Norheim OF, Nowaseb V, Nyakarahuka L, Oh IH, Ohkubo T, Olusanya BO, Omer SB, Opio JN, Orisakwe OE, Pandian JD, Papachristou C, Caicedo AJ, Patten SB, Paul VK, Pavlin BI, Pearce N, Pereira DM, Pervaiz A, Pesudovs K, Petzold M, Pourmalek F, Qato D, Quezada AD, Quistberg DA, Rafay A, Rahimi K, Rahimi-Movaghar V, Ur Rahman S, Raju M, Rana SM, Razavi H, Reilly RQ, Remuzzi G, Richardus JH, Ronfani L, Roy N, Sabin N, Saeedi MY, Sahraian MA, Samonte GM, Sawhney M, Schneider IJ, Schwebel DC, Seedat S, Sepanlou SG, Servan-Mori EE, Sheikhbahaei S, Shibuya K, Shin HH, Shiue I, Shivakoti R, Sigfusdottir ID, Silberberg DH, Silva AP, Simard EP, Singh JA, Skirbekk V, Sliwa K, Soneji S, Soshnikov SS, Sreeramareddy CT, Stathopoulou VK, Stroumpoulis K, Swaminathan S, Sykes BL, Tabb KM, Talongwa RT, Tenkorang EY, Terkawi AS, Thomson AJ, Thorne-Lyman AL, Towbin JA, Traebert J, Tran BX, Dimbuene ZT, Tsilimbaris M, Uchendu US, Ukwaja KN, Uzun SB, Vallely AJ, Vasankari TJ, Venketasubramanian N, Violante FS, Vlassov VV, Vollset SE, Waller S, Wallin MT, Wang L, Wang X, Wang Y, Weichenthal S, Weiderpass E, Weintraub RG, Westerman R, White RA, Wilkinson JD, Williams TN, Woldeyohannes SM, Wong JQ, Xu G, Yang YC, Yano Y, Yentur GK, Yip P, Yonemoto N, Yoon SJ, Younis M, Yu C, Jin KY, El Sayed Zaki M, Zhao Y, Zheng Y, Zhou M, Zhu J, Zou XN, Lopez AD, Vos T. Global, regional, and national incidence and mortality for HIV, tuberculosis, and malaria during 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet 2014;384:1005-1070.
- 3. Breman JG. The ears of the hippopotamus: manifestations, determinants, and estimates of the malaria burden. Am J Trop Med Hyg 2001;64:1-11.
- 4. Cui L, Yan G, Sattabongkot J, Cao Y, Chen B, Chen X, Fan Q, Fang Q, Jongwutiwes S, Parker D, Sirichaisinthop J, Kyaw MP, Su XZ, Yang H, Yang Z, Wang B, Xu J, Zheng B, Zhong D, Zhou G. Malaria in the Greater Mekong Subregion: heterogeneity and complexity. Acta Trop 2012;121:227-239.
- 5. Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM, Artemisinin Resistance in Cambodia 1 (ARC1) Study Consortium. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med 2008;359:2619-2620.
- 6. Phyo AP, Nkhoma S, Stepniewska K, Ashley EA, Nair S, McGready R, ler Moo C, Al-Saai S, Dondorp AM, Lwin KM, Singhasivanon P, Day NP, White NJ, Anderson TJ, Nosten F. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet 2012;379:1960-1966.
- 7. Fairhurst RM, Nayyar GM, Breman JG, Hallett R, Vennerstrom JL, Duong S, Ringwald P, Wellems TE, Plowe CV, Dondorp AM. Artemisinin-resistant malaria: research challenges, opportunities, and public health implications. Am J Trop Med Hyg 2012;87:231-241.
- 8. Ellis RD, Sagara I, Doumbo O, Wu Y. Blood stage vaccines for Plasmodium falciparum: current status and the way forward. Hum Vaccin 2010;6:627-634.
- 9. Miller LH, Ackerman HC, Su XZ, Wellems TE. Malaria biology and disease pathogenesis: insights for new treatments. Nat Med 2013;19:156-167.
- 10. Anders RE, Adda CG, Foley M, Norton RS. Recombinant protein vaccines against the asexual blood stages of Plasmodium falciparum. Hum Vaccin 2010;6:39-53.
- 11. McColl DJ, Silva A, Foley M, Kun JF, Favaloro JM, Thompson JK, Marshall VM, Coppel RL, Kemp DJ, Anders RF. Molecular variation in a novel polymorphic antigen associated with Plasmodium falciparum merozoites. Mol Biochem Parasitol 1994;68:53-67.
- 12. Oeuvray C, Bouharoun-Tayoun H, Grass-Masse H, Lepers JP, Ralamboranto L, Tartar A, Druilhe P. A novel merozoite surface antigen of Plasmodium falciparum (MSP-3) identified by cellular-antibody cooperative mechanism antigenicity and biological activity of antibodies. Mem Inst Oswaldo Cruz 1994;89(Suppl 2):77-80.
- 13. Oeuvray C, Bouharoun-Tayoun H, Gras-Masse H, Bottius E, Kaidoh T, Aikawa M, Filgueira MC, Tartar A, Druilhe P. Merozoite surface protein-3: a malaria protein inducing antibodies that promote Plasmodium falciparum killing by cooperation with blood monocytes. Blood 1994;84:1594-1602.
- 14. Hisaeda H, Saul A, Reece JJ, Kennedy MC, Long CA, Miller LH, Stowers AW. Merozoite surface protein 3 and protection against malaria in Aotus nancymai monkeys. J Infect Dis 2002;185:657-664.
- 15. Carvalho LJ, Oliveira SG, Theisen M, Alves FA, Andrade MC, Zanini GM, Brigido MC, Oeuvray C, Povoa MM, Muniz JA, Druilhe P, Daniel-Ribeiro CT. Immunization of Saimiri sciureus monkeys with Plasmodium falciparum merozoite surface protein-3 and glutamate-rich protein suggests that protection is related to antibody levels. Scand J Immunol 2004;59:363-372.
- 16. Soe S, Theisen M, Roussilhon C, Aye KS, Druilhe P. Association between protection against clinical malaria and antibodies to merozoite surface antigens in an area of hyperendemicity in Myanmar: complementarity between responses to merozoite surface protein 3 and the 220-kilodalton glutamate-rich protein. Infect Immun 2004;72:247-252.
- 17. Polley SD, Tetteh KK, Lloyd JM, Akpogheneta OJ, Greenwood BM, Bojang KA, Conway DJ. Plasmodium falciparum merozoite surface protein 3 is a target of allele-specific immunity and alleles are maintained by natural selection. J Infect Dis 2007;195:279-287.
- 18. Osier FH, Polley SD, Mwangi T, Lowe B, Conway DJ, Marsh K. Naturally acquired antibodies to polymorphic and conserved epitopes of Plasmodium falciparum merozoite surface protein 3. Parasite Immunol 2007;29:387-394.
- 19. Audran R, Cachat M, Lurati F, Soe S, Leroy O, Corradin G, Druilhe P, Spertini F. Phase I malaria vaccine trial with a long synthetic peptide derived from the merozoite surface protein 3 antigen. Infect Immun 2005;73:8017-8026.
- 20. Sirima SB, Nebie I, Ouedraogo A, Tiono AB, Konate AT, Gansane A, Derme AI, Diarra A, Ouedraogo A, Soulama I, Cuzzin-Ouattara N, Cousens S, Leroy O. Safety and immunogenicity of the Plasmodium falciparum merozoite surface protein-3 long synthetic peptide (MSP3-LSP) malaria vaccine in healthy, semi-immune adult males in Burkina Faso, West Africa. Vaccine 2007;25:2723-2732.
- 21. Lusingu JP, Gesase S, Msham S, Francis F, Lemnge M, Seth M, Sembuche S, Rutta A, Minja D, Segeja MD, Bosomprah S, Cousens S, Noor R, Chilengi R, Druilhe P. Satisfactory safety and immunogenicity of MSP3 malaria vaccine candidate in Tanzanian children aged 12-24 months. Malar J 2009;8:163.
- 22. Sirima SB, Tiono AB, Ouedraogo A, Diarra A, Ouedraogo AL, Yaro JB, Ouedraogo E, Gansane A, Bougouma EC, Konate AT, Kaboré Y, Traoré A, Chilengi R, Soulama I, Luty AJ, Druilhe P, Cousens S, Nébié I. Safety and immunogenicity of the malaria vaccine candidate MSP3 long synthetic peptide in 12-24 months-old Burkinabe children. PLoS One 2009;4:e7549.
- 23. McColl DJ, Anders RF. Conservation of structural motifs and antigenic diversity in the Plasmodium falciparum merozoite surface protein-3 (MSP-3). Mol Biochem Parasitol 1997;90:21-31.
- 24. Huber W, Felger I, Matile H, Lipps HJ, Steiger S, Beck HP. Limited sequence polymorphism in the Plasmodium falciparum merozoite surface protein 3. Mol Biochem Parasitol 1997;87:231-234.
- 25. Jordan SJ, Branch OH, Castro JC, Oster RA, Rayner JC. Genetic diversity of the malaria vaccine candidate Plasmodium falciparum merozoite surface protein-3 in a hypoendemic transmission environment. Am J Trop Med Hyg 2009;80:479-486.
- 26. Ebrahimzadeh A, Mohammadi S, Jamshidi A. Allelic forms of merozoite surface protein-3 in Plasmodium falciparum isolates from Southeast of Iran. Jundishapur J Microbiol 2014;7:e9829.
- 27. Soulama I, Bigoga JD, Ndiaye M, Bougouma EC, Quagraine J, Casimiro PN, Stedman TT, Sirima SB. Genetic diversity of polymorphic vaccine candidate antigens (apical membrane antigen-1, merozoite surface protein-3, and erythrocyte binding antigen-175) in Plasmodium falciparum isolates from western and central Africa. Am J Trop Med Hyg 2011;84:276-284.
- 28. Simpalipan P, Pattaradilokrat S, Siripoon N, Seugorn A, Kaewthamasorn M, Butcher RD, Harnyuttanakorn P. Diversity and population structure of Plasmodium falciparum in Thailand based on the spatial and temporal haplotype patterns of the C-terminal 19-kDa domain of merozoite surface protein-1. Malar J 2014;13:54.
- 29. Pumpaibool T, Arnathau C, Durand P, Kanchanakhan N, Siripoon N, Suegorn A, Sitthi-Amorn C, Renaud F, Harnyuttanakorn P. Genetic diversity and population structure of Plasmodium falciparum in Thailand, a low transmission country. Malar J 2009;8:155.
- 30. Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 2002;419:498-511.
- 31. Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 2010;10:564-567.
- 32. Cui L, Yan G, Sattabongkot J, Chen B, Cao Y, Fan Q, Parker D, Sirichaisinthop J, Su XZ, Yang H, Yang Z, Wang B, Zhou G. Challenges and prospects for malaria elimination in the Greater Mekong Subregion. Acta Trop 2012;121:240-245.
- 33. Pattaradilokrat S, Mu J, Awadalla P, Su X. Genome diversity and applications in genetic studies of the human malaria parasites Plasmodium falciparum and Plasmodium vivax. In Carlton JM, Perkins SL, Deitsch KW eds, Malaria Parasites: Comparative Genomics, Evolution and Molecular Biology. Wymondham, UK. Caister Academic Press; 2013, pp 59-90.
- 34. Kuesap J, Chaijaroenkul W, Ketprathum K, Tattiyapong P, Na-Bangchang K. Evolution of genetic polymorphisms of Plasmodium falciparum merozoite surface protein (PfMSP) in Thailand. Korean J Parasitol 2014;52:105-109.
- 35. Rungsihirunrat K, Chaijaroenkul W, Siripoon N, Seugorn A, Na-Bangchang K. Genotyping of polymorphic marker (MSP3alpha and MSP3beta) genes of Plasmodium vivax field isolates from malaria endemic of Thailand. Trop Med Int Health 2011;16:794-801.
- 36. James S, Moehle K, Renard A, Mueller MS, Vogel D, Zurbriggen R, Pluschke G, Robinson JA. Synthesis, solution structure and immune recognition of an epidermal growth factor-like domain from Plasmodium falciparum merozoite surface protein-1. Chem-Biochem 2006;7:1943-1950.
- 37. Pizarro JC, Chitarra V, Verger D, Holm I, Petres S, Dartevelle S, Nato F, Longacre S, Bentley GA. Crystal structure of a Fab complex formed with PfMSP1-19, the C-terminal fragment of merozoite surface protein 1 from Plasmodium falciparum: a malaria vaccine candidate. J Mol Biol 2003;328:1091-1103.
- 38. Kamolratanakul P, Dhanamun B, Lertmaharit S, Seublingwong T, Udomsangpetch R, Thaithong S. Epidemiological studies of malaria at Pong Nam Ron, eastern Thailand. Southeast Asian J Trop Med Public Health 1994;25:425-429.
- 39. Wangroongsarb P, Sudathip P, Satimai W. Characteristics and malaria prevalence of migrant populations in malaria-endemic areas along the Thai-Cambodian border. Southeast Asian J Trop Med Public Health 2012;43:261-269.
- 40. Bang G, Prieur E, Roussilhon C, Druilhe P. Pre-clinical assessment of novel multivalent MSP3 malaria vaccine constructs. PLoS One 2011;6:e28165.