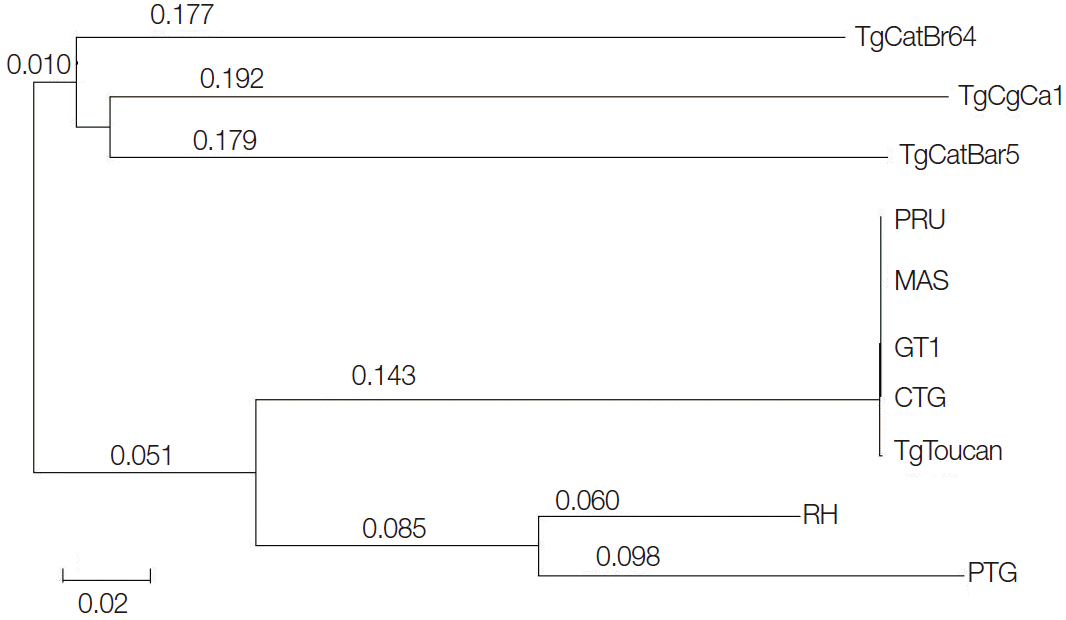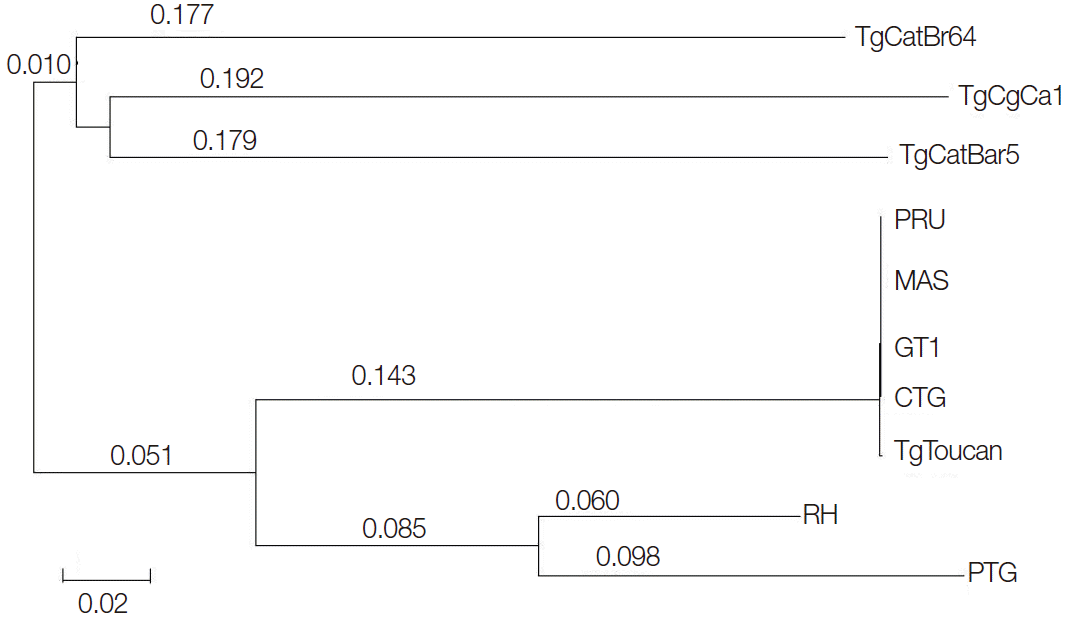Abstract
Toxoplasma gondii, an obligate intracellular protozoan parasite of the phylum Apicomplexa, can infect all warm-blooded vertebrates, including humans, livestock, and marine mammals. The aim of this study was to investigate whether superoxide dismutase (SOD) of T. gondii can be used as a new marker for genetic study or a potential vaccine candidate. The partial genome region of the SOD gene was amplified and sequenced from 10 different T. gondii isolates from different parts of the world, and all the sequences were examined by PCR-RFLP, sequence analysis, and phylogenetic reconstruction. The results showed that partial SOD gene sequences ranged from 1,702 bp to 1,712 bp and A + T contents varied from 50.1% to 51.1% among all examined isolates. Sequence alignment analysis identified total 43 variable nucleotide positions, and these results showed that 97.5% sequence similarity of SOD gene among all examined isolates. Phylogenetic analysis revealed that these SOD sequences were not an effective molecular marker for differential identification of T. gondii strains. The research demonstrated existence of low sequence variation in the SOD gene among T. gondii strains of different genotypes from different hosts and geographical regions.
-
Key words: Toxoplasma gondii, PCR-RFLP, superoxide dismutase, sequence variation, phylogenetic analysis
INTRODUCTION
Toxoplasma gondii, the direct pathogenic factor of toxoplasmosis, is an obligate intracellular protozoan parasite of the phylum Apicomplexa which can infect all warm-blooded vertebrates, including humans, livestock, and marine mammals [
1]. Although most infections are clinically asymptomatic, the parasite can cause severe disease in immunocompromised populations and congenitally infected individuals [
1,
2]. In addition, infections in domestic animals may result in economic losses as well as bring enormous psychological troubles, since it can cause abortion, stillbirth, and neonatal loss [
3].
Unlike many other Apicomplexa parasites which exhibit stronger host specificity,
T. gondii faces vastly numerous hosts and can adapt to various environmental conditions during its complex life cycles, which can be contributed to many different
T. gondii strains and genotypes [
4]. It was popularly believed that
T. gondii had a clonal population structure with 3 predominant lineages, namely types I, II, and III [
5-
7]. Besides these isoforms, it also exists in atypical and recombinant strains [
8,
9]. To better understand the population genetics and molecular epidemiology of this parasite, and to develop more strategies for vaccination, diagnosis, and treatment of toxoplasmosis, it is necessary to study the genetic diversities in
T. gondii [
10,
11].
Superoxide dismutase (SOD), an important enzyme that widely exists in many organisms, including animals, plants, and microorganisms, can promote the conversion of superoxide (O
2−) into hydrogen peroxide and oxygen [
12,
13]. In view of SOD that can eliminate extra superoxide (O
2−) anion in the cells and protect cells from oxidative damages, it has potential applications in medicine, food industry, and agriculture [
12,
14,
15]. In
T. gondii, limited studies have shown that SOD is a typical FeSOD and its activity might be essential for the intracellular growth of both bradyzoite and tachyzoite forms [
16]. To our knowledge, no report has described the sequence variation of SOD gene in different
T. gondii strains. We hereby examined sequence variation of SOD gene among 10
T. gondii isolates from different hosts and geographical regions (different countries), and assess SOD could be used as a new marker for genetic study or a potential vaccine candidate against
T. gondii.
MATERIALS AND METHODS
T. gondii strains
A total of 10 genotyped
T. gondii strains were utilized as shown in
Table 1, and the genomic DNA was prepared as described previously [
17].
The SOD gene was amplified by PCR from genomic DNA of T. gondii with 1 pair of primes: 5´-ATGGTATTCACTTTGCCCCCGCT-3´ (forward prime) and 5´-TCATTTCAAGGCATTCTCCAAG-3´ (reverse primer). The design of primers was based on SOD gene of T. gondii RH isolate available in GenBank database (AF029915). The amplification reaction was performed in a volume of 20 μl containing 2 μl template DNA, 10 μl 2×1 Step buffer (0.5 U Taq polymerase), 1 μl of each primer, and 6 μl RNase-free dH2O. The target DNA was amplified under the following conditions: 94˚C for 30 sec, 63.6˚C for 1 min, and 72˚C for 1 min. The PCR amplification products were confirmed by electrophoresis in a 1.5% agarose gels and staining with ethidium bromide followed by visualization under UV.
The analysis of PCR-RFLP
The SOD PCR amplification products from representative T. gondii strains were digested with restriction enzymes Xba I and EcoR I, respectively, and incubated at 37˚C for 3 hr. The restriction fragments were separated by electrophoresis in 1.5% agarose gel and staining with ethidium bromide, followed by visualization under UV.
Sequence analysis and reconstruction of phylogenetic relationships
The SOD PCR products were purified with the DNA purification kit (TransGen Biotech, Beijing, China) and ligated with the pEASY-T1 vector (TransGen Biotech) according to the manufacturer's instructions, and then transformed into
Escherichia coli DH5α competent cells. The transformed cells carrying the insert were successively selected by blue-white screening, PCR, and restriction enzyme digestion. The positive colonies were sequenced by Beijing Genomics Institute Company (Beijing, China). The obtained SOD gene sequences from different
T. gondii strains were aligned using Multiple Sequence Alignment Program Clustal ×1.83. Phylogenetic reconstructions based on the sequences of SOD gene among different
T. gondii strains were performed using 3 methodologies, namely maximum parsimony (MP), Bayesian inference (BI), and maximum likelihood (ML) [
18,
19].
RESULTS
PCR-RFLP and sequence analysis
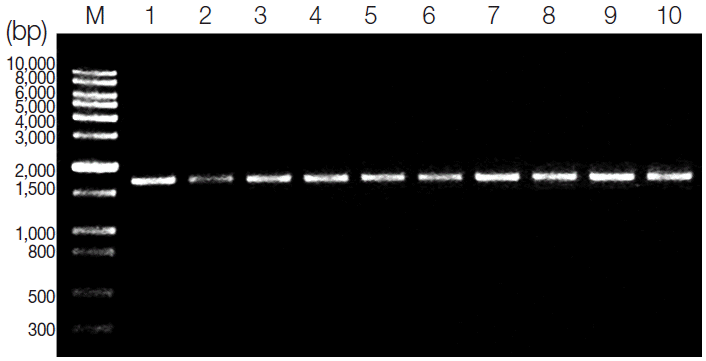
The amplification of the SOD gene resulted in a single product of approximate 1,700 bp in length on agarose gel for all tested
T. gondii strains (
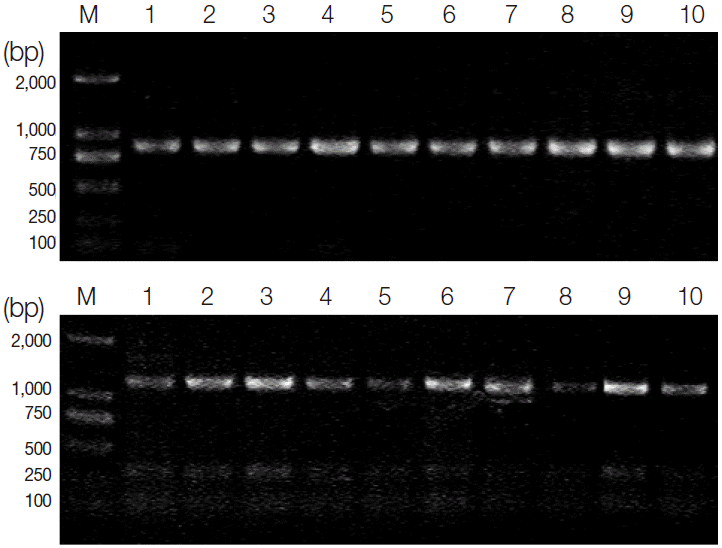
Fig. 1). There are no obvious differences in all tested
T. gondii strains after digestion of the amplified SOD products with
EcoR I and
Xba I, revealing that subtypes I, II, and III could not be differentiated in this condition (
Fig. 2). Then, the amplicons of all isolates were sequenced. The sequencing results showed that the SOD gene was 1,712 bp in length for the TgCatBr64, 1,709 bp in length for PTG, 1,706 bp in length for RH, 1,702 bp in length for TgCgCa1, and 1,707 bp for the remaining 6 strains. Besides, the A + T contents of these sequences ranged from 50.1% to 51.1% among all isolates. The alignment of all 10 sequences revealed nucleotide polymorphisms at 43 positions, with an intraspecific variation of 0-1.0% (
Table 2). Of these variable nucleotide positions, there were 29 transitions (C↔T, T↔G, A↔C, and A↔G), 7 transversions (A↔T and C↔G), and 7 deletions in all the sequences. These results showed 97.5% sequence similarity of SOD gene among all the tested isolates, suggesting considerable sequence diversity. Amino acid polymorphisms at 24 positions were detected among 10
T. gondii strains, including 20 amino acid position substitutions and 4 amino acid position deletions (
Table 3).
Phylogenetic analysis of the 10 examined
T. gondii isolates based on the SOD gene sequences demonstrated 2 major clusters (
Fig. 3). TgCatBr5, TgCatBr64, and TgCgCa1 strains were clustered in 1 clade, and PRU, MAS, GT1, CTG, TgTgucan, PTG, and RH were clustered in the other clade. Overall, the topologies of all trees based on nucleotide sequences inferred by 2 different methods were similar, with only small differences of bootstrap values.
DISCUSSION
SOD widely exists in many organisms and plays a crucial role in eliminating the extra superoxide anion (O
2−) in the cells to avoid oxidative damages [
12-
15]. In
T. gondii, SOD is an iron-containing type, which correlates with the protection of the parasite [
16]. However, SOD enzymes from C and RH strain tachyzoites do not appear to be the basis for differences in virulence to mice [
20]. In the present study, we cloned and sequenced the partial genome sequence of SOD gene among 10
T. gondii isolates from different hosts and geographical regions and examined genetic diversity of SOD locus by the techniques of PCR-RFLP, sequence analysis, and phylogenetic reconstruction. The results revealed nucleotide polymorphisms at 43 positions and amino acid polymorphisms at 24 positions, suggesting low sequence variability among all the tested isolates.
PCR-RFLP has been widely used in analysis of specific genetic loci for
T. gondii genotyping. Multilocus PCR-RFLP marker is a high resolution for identification of parasites, although it requires a huge investment of time to test and optimize each marker [
11]. By contrast, single PCR-RFLP marker is simply and more convenient. Then, studies on single marker loci (e.g. GRA5, GRA6, and ROP17) have shown signs of positive selection, and could be sufficient for genotyping of
T. gondii isolates. [
21-
23].
In our study, however, 3 clonal lineages (types I, II, and III) cannot be differentiated in 10 examined
T. gondii strains using a single PCR-RFLP marker SOD. One possible reason is that the sequence variation of SOD could not be located in restriction enzyme sites, to some degree, leading to loss of many polymorphisms in this situation [
24].
The direct sequencing of genomic regions can detect small deletions and insertions (e.g., indels) and single nucleotide polymorphisms (SNPs) in the genomic regions, and hence it is capable of testing more genetic diversity compared with PCR-RFLP. Based on the partial sequences of the SOD locus, phylogenetic analysis revealed 2 major clusters with only a little difference of bootstrap values, and it cannot differentiate
T. gondii strains to their genotypes, implying a low genetic diversity. Our results were similar to some studies such as MIC13, eIF4A [
25,
26] and different from other genetic markers including GRA5, GRA6, and ROP17 [
21-
23]. The low sequence variation (0-1.0%) in the partial SOD gene suggests that SOD gene could not be an ideal genetic marker for differentiation of the
T. gondii strains or intraspecific phylogenetic analyses.
Notes
-
We have no conflict of interest related to this work.
This study was financially supported by the National Natural Science Foundation of China (grant no. 81471974 and 81171604). Dr. Chunlei Su at Department of Microbiology, The University of Tennessee, Knoxville, USA is thanked for providing T. gondii reference strains.
Fig. 1.Analysis of PCR products of SOD gene from 10 examined T. gondii strains using electrophoresis. Lane M; DNA size marker 10,000. Lanes 1-10; T. gondii type I (GT1, RH), type II (PTG, PRU), type III (CTG) strains and other strains (TgCgCa1, MAS ,TgCatBr5 ,TgCatBr64, and TgToucan), respectively.
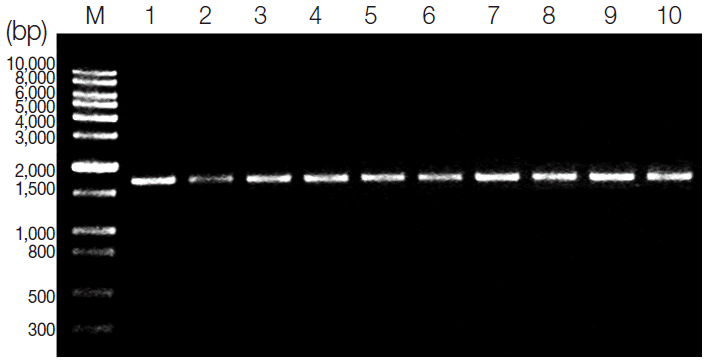
Fig. 2.PCR-RFLP analysis of SOD genes using restriction endonucleases EcoR I (upper) and Xba I (bottom). Lane M; DNA size marker 2,000. Lanes 1-10; T. gondii type I (GT1, RH), type II (PTG, PRU), type III (CTG) strains and other strains (TgCgCa1, MAS, TgCatBr5, TgCatBr64, and TgToucan), respectively.
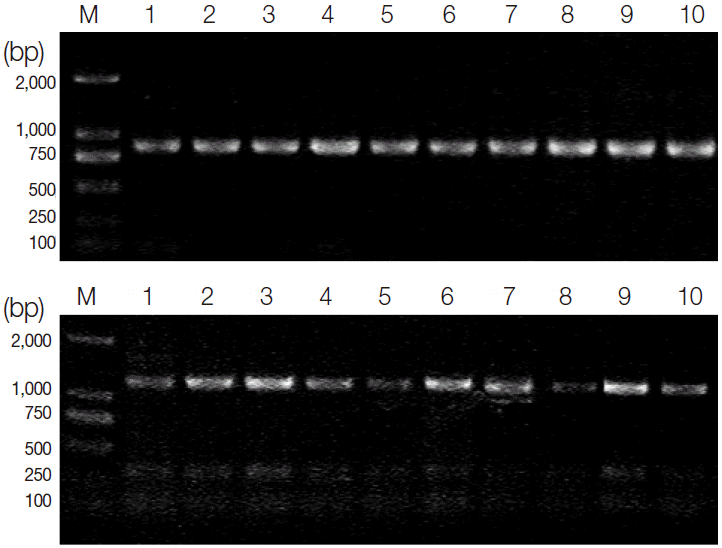
Fig. 3.Phylogenetic analyses of 10 Toxoplasma gondii strains based on analysis of the SOD gene sequences. The tree was built by Bayesian inference (BI) analysis. The numbers along branches indicate bootstrap values resulting from BI analysis.
Table 1.
Toxoplasma gondii strains subjected to SOD gene sequence analysis
Table 1.
|
Strain |
Host |
Geographical origin |
Genotypea
|
|
RH |
Human |
France |
Reference, Type I, ToxoDB #10 |
|
GT1 |
Goat |
United States |
Reference, Type I, ToxoDB #10 |
|
PTG |
Sheep |
United States |
Reference, Type II, ToxoDB #1 |
|
Prugniaud (PRU) |
Human |
France |
Reference, Type II, ToxoDB #1 |
|
CTG |
Cat |
United States |
Reference, Type III, ToxoDB #2 |
|
TgCgCa1 |
Cougar |
Canada |
Reference, ToxoDB #66 |
|
MAS |
Human |
France |
Reference, ToxoDB #17 |
|
TgCatBr5 |
Cat |
Brazil |
Reference ToxoDB #19 |
|
TgCatBr64 |
Cat |
Brazil |
Reference ToxoDB #111 |
|
TgToucan (TgRsCr1) |
Toucan |
Costa Rica |
Reference, ToxoDB #52 |
Table 2.Nucleotide polymorphisms of the SOD gene coding region within Toxoplasma gondii strains
Table 2.
|
RH |
CTG |
GT1 |
MAS |
PRU |
PTG |
TgCatBr5 |
TgCatBr64 |
TgCgCa1 |
TgToucan |
|
1403 |
A |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
C |
﹡ |
﹡ |
|
1437 |
A |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
T |
﹡ |
﹡ |
|
1480 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
C |
﹡ |
﹡ |
|
1501 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
- |
﹡ |
- |
﹡ |
|
1503 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
T |
﹡ |
﹡ |
﹡ |
|
1504 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
T |
﹡ |
|
1518 |
A |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
T |
﹡ |
﹡ |
﹡ |
|
1519 |
C |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
A |
﹡ |
﹡ |
﹡ |
|
1520 |
T |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
C |
﹡ |
﹡ |
﹡ |
|
1521 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
T |
﹡ |
﹡ |
﹡ |
|
1532 |
C |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
T |
﹡ |
﹡ |
﹡ |
|
1535 |
C |
﹡ |
﹡ |
﹡ |
﹡ |
T |
﹡ |
﹡ |
﹡ |
﹡ |
|
1545 |
A |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
G |
﹡ |
|
1547 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
C |
﹡ |
﹡ |
|
1549 |
A |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
G |
﹡ |
|
1555 |
C |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
T |
﹡ |
﹡ |
﹡ |
|
1556 |
A |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
T |
﹡ |
﹡ |
﹡ |
|
1557 |
T |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
C |
﹡ |
﹡ |
﹡ |
|
1558 |
C |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
T |
﹡ |
﹡ |
﹡ |
|
1566 |
A |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
G |
﹡ |
﹡ |
﹡ |
|
1567 |
A |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
T |
﹡ |
|
1568 |
C |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
T |
﹡ |
|
1574 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
A |
﹡ |
﹡ |
﹡ |
﹡ |
|
1575 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
A |
﹡ |
﹡ |
﹡ |
|
1584 |
T |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
C |
﹡ |
﹡ |
|
1585 |
C |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
- |
﹡ |
﹡ |
|
1586 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
- |
﹡ |
﹡ |
|
1587 |
A |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
G |
﹡ |
﹡ |
|
1590 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
A |
A |
﹡ |
﹡ |
﹡ |
|
1599 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
A |
﹡ |
|
1601 |
C |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
- |
﹡ |
|
1614 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
A |
﹡ |
﹡ |
﹡ |
﹡ |
|
1616 |
T |
﹡ |
﹡ |
﹡ |
﹡ |
G |
﹡ |
﹡ |
﹡ |
﹡ |
|
1618 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
T |
﹡ |
﹡ |
﹡ |
﹡ |
|
1620 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
A |
﹡ |
|
1628 |
T |
﹡ |
﹡ |
﹡ |
﹡ |
A |
﹡ |
﹡ |
﹡ |
﹡ |
|
1635 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
C |
|
1637 |
C |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
- |
﹡ |
﹡ |
﹡ |
|
1668 |
A |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
- |
﹡ |
|
1669 |
C |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
A |
﹡ |
﹡ |
﹡ |
|
1679 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
A |
﹡ |
﹡ |
﹡ |
|
1685 |
C |
﹡ |
﹡ |
﹡ |
﹡ |
A |
﹡ |
﹡ |
﹡ |
﹡ |
|
1793 |
C |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
- |
﹡ |
﹡ |
﹡ |
Table 3.Amino acid changes of the SOD gene coding region among ten Toxoplasma gondii strains
Table 3.
|
RH |
CTG |
GT1 |
MAS |
PRU |
PTG |
TgCatBr5 |
TgCatBr64 |
TgCgCa1 |
TgToucan |
|
66 |
N |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
Y |
﹡ |
﹡ |
|
80 |
S |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
T |
﹡ |
﹡ |
|
87 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
- |
﹡ |
- |
﹡ |
|
88 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
C |
﹡ |
V |
﹡ |
|
93 |
T |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
S |
﹡ |
﹡ |
|
94 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
W |
﹡ |
﹡ |
|
102 |
K |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
N |
﹡ |
E |
﹡ |
|
103 |
E |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
G |
﹡ |
|
105 |
T |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
I |
﹡ |
﹡ |
﹡ |
|
106 |
S |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
F |
﹡ |
﹡ |
﹡ |
|
109 |
N |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
I |
﹡ |
|
112 |
D |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
N |
﹡ |
﹡ |
﹡ |
|
115 |
S |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
P |
﹡ |
﹡ |
|
116 |
K |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
E |
﹡ |
﹡ |
|
117 |
V |
﹡ |
﹡ |
﹡ |
﹡ |
I |
I |
﹡ |
﹡ |
﹡ |
|
120 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
S |
﹡ |
|
125 |
G |
﹡ |
﹡ |
﹡ |
﹡ |
S |
﹡ |
﹡ |
﹡ |
﹡ |
|
126 |
W |
﹡ |
﹡ |
﹡ |
﹡ |
F |
﹡ |
﹡ |
﹡ |
﹡ |
|
127 |
A |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
T |
﹡ |
|
129 |
L |
﹡ |
﹡ |
﹡ |
﹡ |
H |
﹡ |
﹡ |
﹡ |
﹡ |
|
132 |
D |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
- |
﹡ |
﹡ |
H |
|
143 |
T |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
N |
﹡ |
- |
﹡ |
|
148 |
T |
﹡ |
﹡ |
﹡ |
﹡ |
N |
﹡ |
﹡ |
﹡ |
﹡ |
|
151 |
T |
﹡ |
﹡ |
﹡ |
﹡ |
﹡ |
- |
﹡ |
﹡ |
﹡ |
References
- 1. Montoya JG, Liesenfeld O. Toxoplasmosis. Lancet 2004;363:1965-1976.
- 2. Weiss LM, Dubey JP. Toxoplasmosis: a history of clinical observations. Int J Parasitol 2009;39:895-901.
- 3. Cenci-Goga BT, Rossitto PV, Sechi P, McCrindle CM, Cullor JS. Toxoplasma in animals, food, and humans: an old parasite of new concern. Foodborne Pathog 2011;8:751-762.
- 4. Sibley LD, Ajioka JW. Population structure of Toxoplasma gondii: clonal expansion driven by infrequent recombination and selective sweeps. Annu Rev Microbiol 2008;62:329-351.
- 5. Sibley LD, Boothroyd JC. Virulent strains of Toxoplasma gondii comprise a single clonal lineage. Nature 1992;359:82-85.
- 6. Howe DK, Sibley LD. Toxoplasma gondii comprises three clonal lineages: correlation of parasite genotype with human disease. J Infect Dis 1995;172:1561-1566.
- 7. Sibley LD, Mordue DG, Su CL, Robben PM, Howe DK. Genetic approaches to studying virulence and pathogenesis in Toxoplasma gondii. Philos Trans Roy Soc B 2002;357:81-88.
- 8. Dardé ML. Toxoplasma gondii, “new” genotypes and virulence. Parasite 2008;15:366-371.
- 9. Delhaes L, Ajzenberg D, Sicot B, Bourgeot P, Dardé ML, Dei-Cas E, Houfflin-Debarge V. Severe congenital toxoplasmosis due to a Toxoplasma gondii strain with an atypical genotype: case report and review. Prenat Diagn 2010;30:902-905.
- 10. Robert-Gangneux F, Dardé ML. Epidemiology of and diagnostic strategies for toxoplasmosis. Clin Microbiol Rev 2012;25:264-296.
- 11. Su C, Zhang X, Dubey JP. Genotyping of Toxoplasma gondii by multilocus PCR-RFLP markers: a high resolution and simple method for identification of parasites. Int J Parasitol 2006;36:841-848.
- 12. Miller AF. Superoxide dismutase: ancient enzymes and new insights. FEBS Lett 2012;586:585-595.
- 13. Fukai T, Ushio-Fukai M. Superoxide dismutase: role in redox signaling, vascular function, and diseases. Antioxid Redox Signal 2011;15:1583-606.
- 14. Miao L, St Clair DK. Regulation of superoxide dismutase genes: implications in disease. Free Radic Biol Med 2009;47:344-356.
- 15. Noor R, Mittal S, Iqbal J. Superoxide dismutase-applications and relevance to human diseases. Med Sci Monit 2002;8:210-215.
- 16. Odberg-Ferragut C1, Renault JP, Viscogliosi E, Toursel C, Briche I, Engels A, Lepage G, Morgenstern-Badarau I, Camus D, Tomavo S, Dive D. Molecular cloning, expression analysis and iron metal cofactor characterisation of a superoxide dismutase from Toxoplasma gondii. Mol Biochem Parasitol 2000;106:121-129.
- 17. Su C, Shwab EK, Zhou P, Zhu XQ, Dubey JP. Moving towards an integrated approach to molecular detection and identification of Toxoplasma gondii. Parasitology 2010;137:1-11.
- 18. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997;24:4876-4882.
- 19. Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003;19:1572-1574.
- 20. Sibley LD, Lawson R, Weidner E. Superoxide dismutase and catalase in Toxoplasma gondii. Mol Biochem Parasitol 1986;19:83-87.
- 21. Chen J, Li ZY, Zhou DH, Liu GH, Zhu XQ. Genetic diversity among Toxoplasma gondii strains from different hosts and geographical regions revealed by sequence analysis of GRA5 gene. Parasit Vectors 2012;5:279.
- 22. Fazaeli A, Carter PE, Darde ML, Pennington TH. Molecular typing of Toxoplasma gondii strains by GRA6 gene sequence analysis. Int J Parasitol 2000;30:637-642.
- 23. Zhang NZ, Xu Y, Huang SY, Zhou DH, Wang RA, Zhu XQ. Sequence variation in Toxoplasma gondii rop17 gene among strains from different hosts and geographical locations. ScientificWorldJournal 2014;2014:349325.
- 24. Sibley LD, Khan A, Ajioka JW, Rosenthal BM. Genetic diversity of Toxoplasma gondii in animals and humans. Philos Trans Roy Soc Lond B Biol Sci 2009;364:2749-2761.
- 25. Ren D, Zhou DH, Xu MJ, Zhou Y. Sequence variation in Toxoplasma gondii MIC13 gene among isolates from different hosts and geographical locations. Afr J Microbiol Res 2012;6:1333-1337.
- 26. Chen J, Fang SF, Zhou DH, Li ZY, Liu GH, Zhu XQ. Sequence variation in the Toxoplasma gondii eIF4A gene among strains from different hosts and geographical locations. Genet Mol Res 2014;13:3356-3361.